install.packages("palmerpenguins")Demo 01: Into the tidyverse
What is Exploratory Data Analysis (EDA)?
(Broadly speaking) EDA = questions about data + wrangling + visualization
R for Data Science: “EDA is a state of mind”, an iterative cycle:
generate questions
answer via transformations and visualizations
Example of questions?
What type of variation do the variables display?
What type of relationships exist between variables?
Goal: develop understanding and become familiar with your data
EDA is NOT a replacement for statistical inference and learning
EDA is an important and necessary step to build intuition
We tackle the challenges of EDA with a data science workflow. An example of this according to Hadley Wickham in R for Data Science:
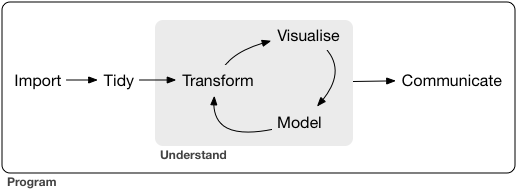
Aspects of data wrangling:
import: reading in data (e.g.,
read_csv())tidy: rows = observations, columns = variables (i.e. tabular data)
transform: filter observations, create new variables, summarize, etc.
Working with penguins
In R, there are many libraries or packages/groups of programs that are not permanently stored in R, so we have to load them when we want to use them. You can load an R package by typing library(package_name). (Sometimes we need to download/install the package first, as described in HW0.)
Throughout this demo we will use the palmerpenguins dataset. To access the data, you will need to install the palmerpenguins package:
Import the penguins dataset by loading the palmerpenguins package using the library function and then access the data with the data() function:
library(palmerpenguins)
data(penguins)View some basic info about the penguins dataset:
# displays same info as c(nrow(penguins), ncol(penguins))
dim(penguins) [1] 344 8class(penguins)[1] "tbl_df" "tbl" "data.frame"tbl (pronounced tibble) is the tidyverse way of storing tabular data, like a spreadsheet or data.frame
I assure you that you’ll run into errors as you code in R; in fact, my attitude as a coder is that something is wrong if I never get any errors while working on a project. When you run into an error, your first reaction may be to panic and post a question to Piazza. However, checking help documentation in R can be a great way to figure out what’s going wrong. (For good or bad, I end up having to read help documentation almost every day of my life - because, well, I regularly make mistakes in R.)
Look at the help documentation for penguins by typing help(penguins) in the Console. What are the names of the variables in this dataset? How many observations are in this dataset?
help(penguins)You should always look at your data before doing anything: view the first 6 (by default) rows with head()
head(penguins) # Try just typing penguins into your console, what happens?# A tibble: 6 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
# ℹ 2 more variables: sex <fct>, year <int>Is our penguins dataset tidy?
Each row = a single penguin
Each column = different measurement about the penguins (can print out column names directly with
colnames(penguins)ornames(penguins))
We’ll now explore differences among the penguins using the tidyverse.
Let the data wrangling begin…
First, load the tidyverse for exploring the data - and do NOT worry about the warning messages that will pop-up! Warning messages will tell you when other packages that are loaded may have functions replaced with the most recent package you’ve loaded. In general though, you should just be concerned when an error message pops up (errors are different than warnings!).
library(tidyverse)Warning: package 'ggplot2' was built under R version 4.2.3Warning: package 'tidyr' was built under R version 4.2.3Warning: package 'readr' was built under R version 4.2.3Warning: package 'dplyr' was built under R version 4.2.3Warning: package 'stringr' was built under R version 4.2.3── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.0 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsWe’ll start by summarizing continuous (e.g., bill_length_mm, flipper_length_mm) and categorical (e.g., species, island) variables in different ways.
We can compute summary statistics for continuous variables with the summary() function:
summary(penguins$bill_length_mm) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
32.10 39.23 44.45 43.92 48.50 59.60 2 Compute counts of categorical variables with table() function:
table("island" = penguins$island) # be careful it ignores NA values!island
Biscoe Dream Torgersen
168 124 52 How do we remove the penguins with missing bill_length_mm values? Within the tidyverse, dplyr is a package with functions for data wrangling (because it’s within the tidyverse that means you do NOT have to load it separately with library(dplyr) after using library(tidyverse)!). It’s considered a “grammar of data manipulation”: dplyr functions are verbs, datasets are nouns.
We can filter() our dataset to choose observations meeting conditions:
clean_penguins <- filter(penguins, !is.na(bill_length_mm))
# Use help(is.na) to see what it returns. And then observe
# that the ! operator means to negate what comes after it.
# This means !TRUE == FALSE (i.e., opposite of TRUE is equal to FALSE).
nrow(penguins) - nrow(clean_penguins) # Difference in rows[1] 2If we want to only consider a subset of columns in our data, we can select() variables of interest:
sel_penguins <- select(clean_penguins, species, island, bill_length_mm, flipper_length_mm)
head(sel_penguins, n = 3)# A tibble: 3 × 4
species island bill_length_mm flipper_length_mm
<fct> <fct> <dbl> <int>
1 Adelie Torgersen 39.1 181
2 Adelie Torgersen 39.5 186
3 Adelie Torgersen 40.3 195We can arrange() our dataset to sort observations by variables:
bill_penguins <- arrange(sel_penguins, desc(bill_length_mm)) # use desc() for descending order
head(bill_penguins, n = 3)# A tibble: 3 × 4
species island bill_length_mm flipper_length_mm
<fct> <fct> <dbl> <int>
1 Gentoo Biscoe 59.6 230
2 Chinstrap Dream 58 181
3 Gentoo Biscoe 55.9 228We can summarize() our dataset to one row based on functions of variables:
summarize(bill_penguins, max(bill_length_mm), median(flipper_length_mm))# A tibble: 1 × 2
`max(bill_length_mm)` `median(flipper_length_mm)`
<dbl> <dbl>
1 59.6 197We can mutate() our dataset to create new variables:
new_penguins <- mutate(bill_penguins,
bill_flipper_ratio = bill_length_mm / flipper_length_mm,
flipper_bill_ratio = flipper_length_mm / bill_length_mm)
head(new_penguins, n = 1)# A tibble: 1 × 6
species island bill_length_mm flipper_length_mm bill_flipper_ratio
<fct> <fct> <dbl> <int> <dbl>
1 Gentoo Biscoe 59.6 230 0.259
# ℹ 1 more variable: flipper_bill_ratio <dbl>How do we perform several of these actions?
head(arrange(select(mutate(filter(penguins, !is.na(flipper_length_mm)), bill_flipper_ratio = bill_length_mm / flipper_length_mm), species, island, bill_flipper_ratio), desc(bill_flipper_ratio)), n = 1)# A tibble: 1 × 3
species island bill_flipper_ratio
<fct> <fct> <dbl>
1 Chinstrap Dream 0.320That’s awfully annoying to do, and also difficult to read…
Enter the pipeline
The |> (pipe) operator is used in the to chain commands together. Note: you can also use the tidyverse pipe %>% (from magrittr), but |> is the built-in pipe that is native to new versions of R without loading the tidyverse.
|> directs the data analyis pipeline: output of one function pipes into input of the next function
penguins |>
filter(!is.na(flipper_length_mm)) |>
mutate(bill_flipper_ratio = bill_length_mm / flipper_length_mm) |>
select(species, island, bill_flipper_ratio) |>
arrange(desc(bill_flipper_ratio)) |>
head(n = 5)# A tibble: 5 × 3
species island bill_flipper_ratio
<fct> <fct> <dbl>
1 Chinstrap Dream 0.320
2 Chinstrap Dream 0.275
3 Chinstrap Dream 0.270
4 Chinstrap Dream 0.270
5 Chinstrap Dream 0.268More pipeline actions!
Instead of head(), we can slice() our dataset to choose the observations based on the position
penguins |>
filter(!is.na(flipper_length_mm)) |>
mutate(bill_flipper_ratio = bill_length_mm / flipper_length_mm) |>
select(species, island, bill_flipper_ratio) |>
arrange(desc(bill_flipper_ratio)) |>
slice(c(1, 2, 10, 100))# A tibble: 4 × 3
species island bill_flipper_ratio
<fct> <fct> <dbl>
1 Chinstrap Dream 0.320
2 Chinstrap Dream 0.275
3 Chinstrap Dream 0.264
4 Gentoo Biscoe 0.227Grouped operations
We group_by() to split our dataset into groups based on a variable’s values
penguins |>
filter(!is.na(flipper_length_mm)) |>
group_by(island) |>
summarize(n_penguins = n(), #counts number of rows in group
ave_flipper_length = mean(flipper_length_mm),
sum_bill_depth = sum(bill_depth_mm),
.groups = "drop") |> # all levels of grouping dropping
arrange(desc(n_penguins)) |>
slice(1:5)# A tibble: 3 × 4
island n_penguins ave_flipper_length sum_bill_depth
<fct> <int> <dbl> <dbl>
1 Biscoe 167 210. 2651.
2 Dream 124 193. 2275.
3 Torgersen 51 191. 940.group_by()is only useful in a pipeline (e.g. withsummarize()), and pay attention to its behaviorspecify the
.groupsfield to decide if observations remain grouped or not after summarizing (you can also useungroup()for this as well)
Putting it all together…
As your own exercise, create a tidy dataset where each row == an island with the following variables:
- number of penguins,
- number of unique species on the island (see
help(unique)), - average
body_mass_g, - variance (see
help(var)) ofbill_depth_mm
Prior to making those variables, make sure to filter missings and also only consider female penguins. Then arrange the islands in order of the average body_mass_g:
# INSERT YOUR CODE HERE