[1] Adelie Adelie Adelie Adelie Adelie Adelie
Levels: Adelie Chinstrap GentooPrinciples and Visualizations for 1D Categorical Data
2024-08-28
Reminders, previously, and today…
HW1 was UPDATED and is due next Wednesday - complete the GenAI Literacy module ON TIME!
Complete HW0 by Thursday night! Confirms you have everything installed and can render
.qmdfiles to PDF viatinytex
Walked through course logistics (READ THE SYLLABUS)
Introduced the Grammar of Graphics and
ggplot2basics
TODAY:
Discuss data visualization principles and the role of infographics
Visualizing categorical data (starting with 1D)
In the beginning…
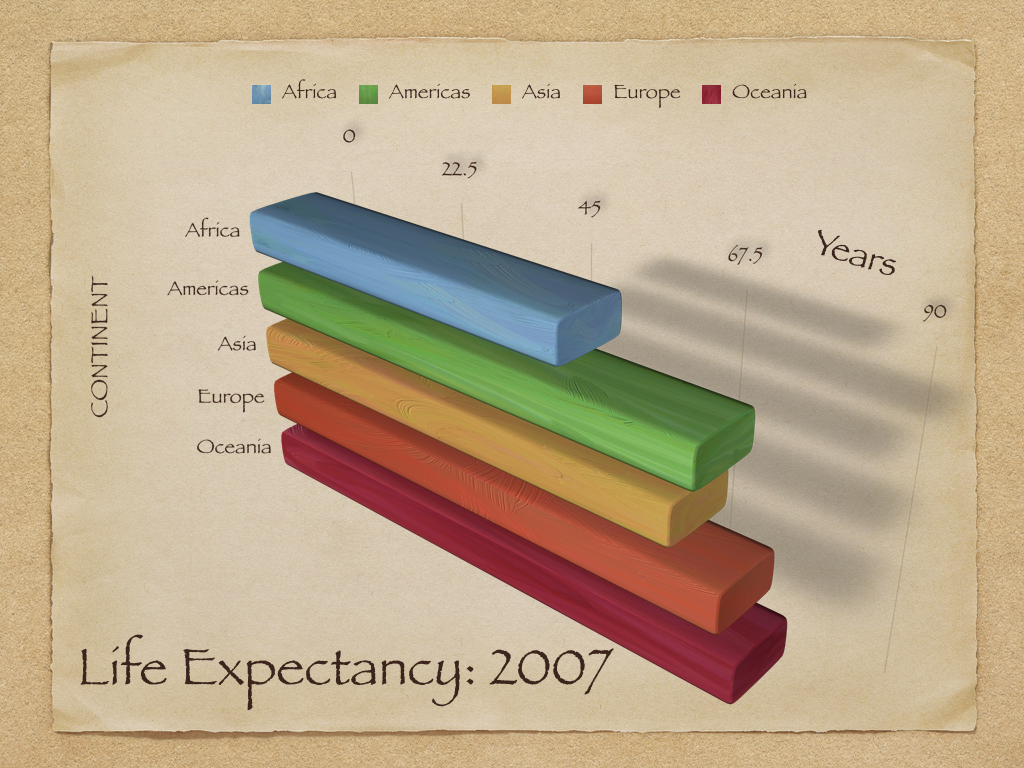
Michael Florent van Langren published the first (known) statistical graphic in 1644

Plots different estimates of the longitudinal distance between Toledo, Spain and Rome, Italy
i.e., visualization of collected data to aid in estimation of parameter

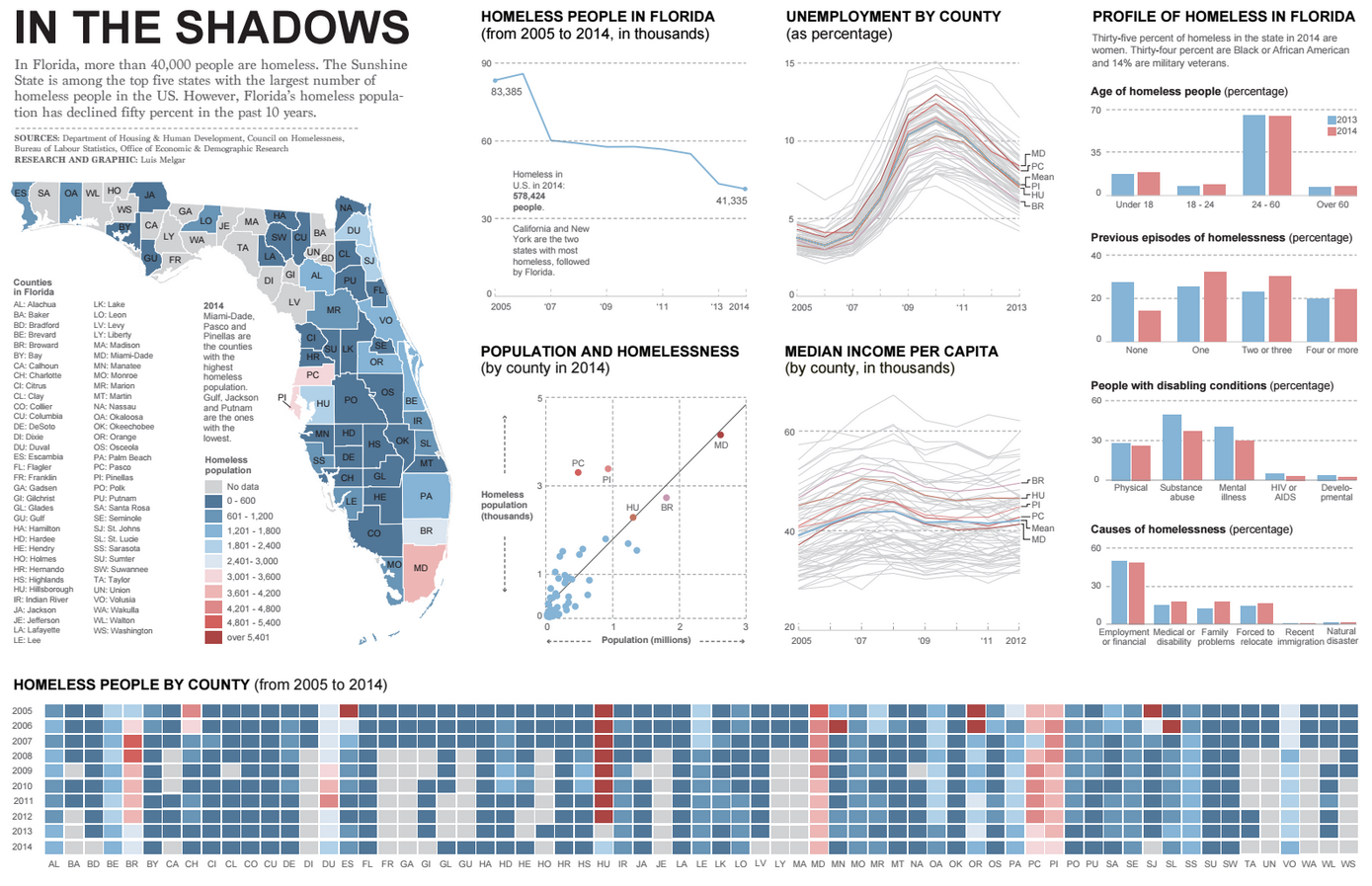
John Snow Knows Something About Cholera
Charles Minard’s Map of Napoleon’s Russian Disaster
Florence Nightingale’s Rose Diagram
Milestones in Data Visualization History
Edward Tufte’s Principles of Data Visualization
Graphics: visually display measured quantities by combining points, lines, coordinate systems, numbers, symbols, words, shading, color
Often our goal is to show data and/or communicate a story
Induce viewer to think about substance, not graphical methodology
Make large, complex datasets more coherent
Encourage comparison of different pieces of data
Describe, explore, and identify relationships
Avoid data distortion and data decoration
Use consistent graph design
Avoid graphs that lead to misleading conclusions!
How to Fail this Class:
What about this spiral?

Infographics to communicate a story (check out FlowingData for more examples)

Alberto Cairo and the art of insight
1D Categorical Data
Two different versions of categorical:
- Nominal: coded with arbitrary numbers, i.e., no real order
- Examples: race, gender, species, text
- Ordinal: levels with a meaningful order
- Examples: education level, grades, ranks
NOTE: R and ggplot considers a categorical variable to be factor
Rwill always treat categorical variables as ordinal! Defaults to alphabetical…We will need to manually define the
factorlevels
1D categorical data structure
Observations are collected into a vector \((x_1, \dots, x_n)\), where \(n\) is number of observations
Each observed value \(x_i\) can only belong to one category level \(\{ C_1, C_2, \dots \}\)
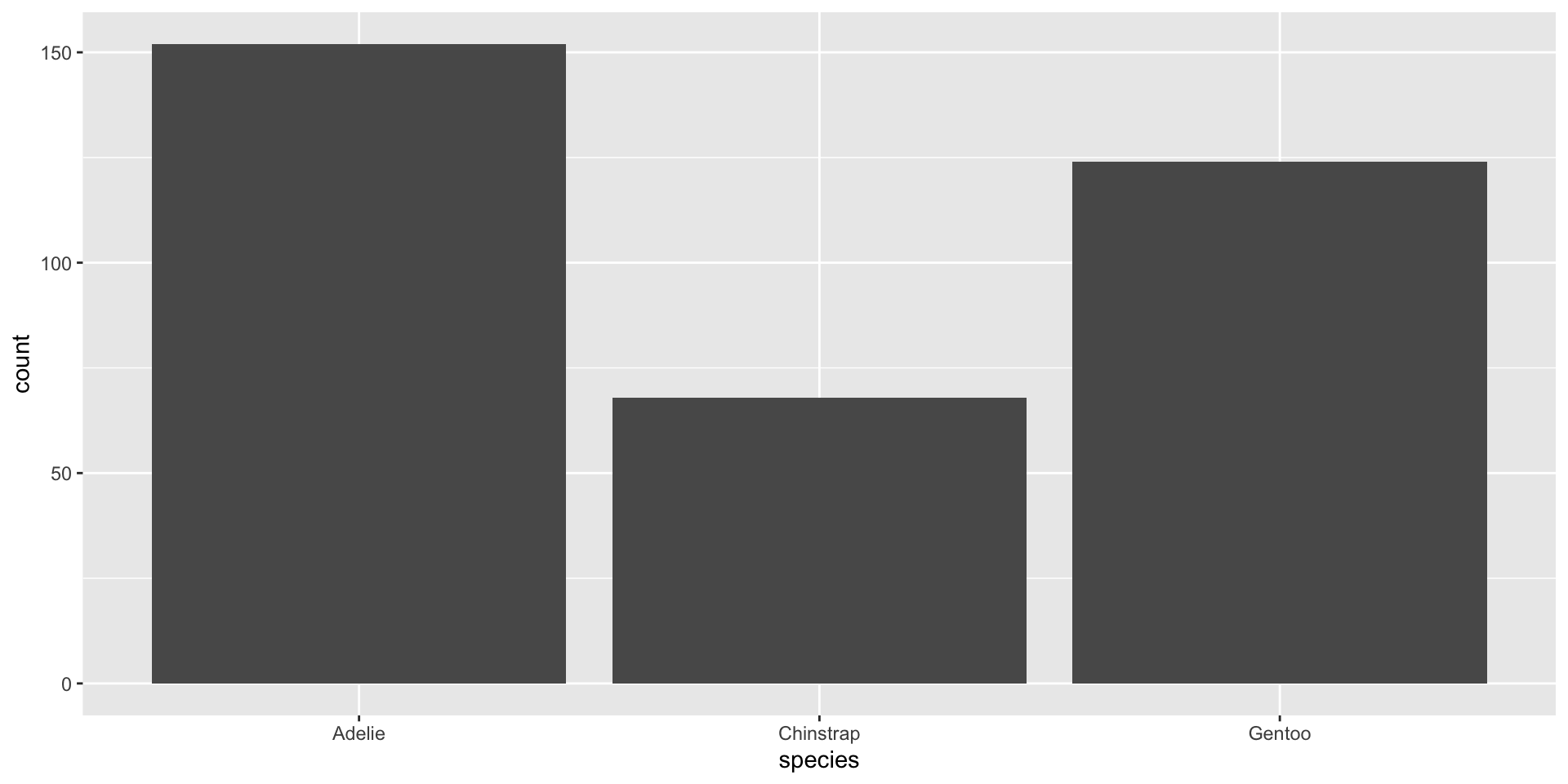
Look at penguins data from the palmerpenguins package, focusing on species:
How could we summarize these data? What information would you report?
Area plots
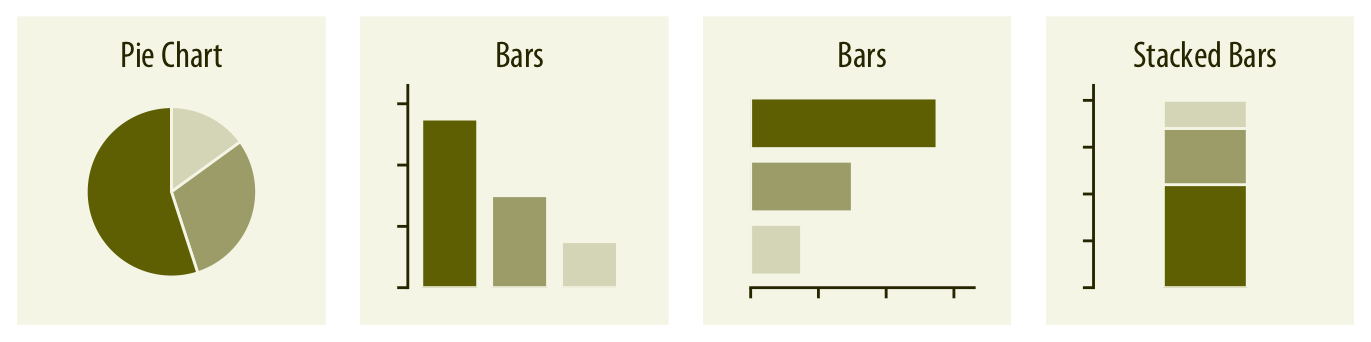
Each area corresponds to one categorical level
Area is proportional to counts/frequencies/percentages
Differences between areas correspond to differences between counts/frequencies/percentages
Bar charts
Behind the scenes: statistical summaries
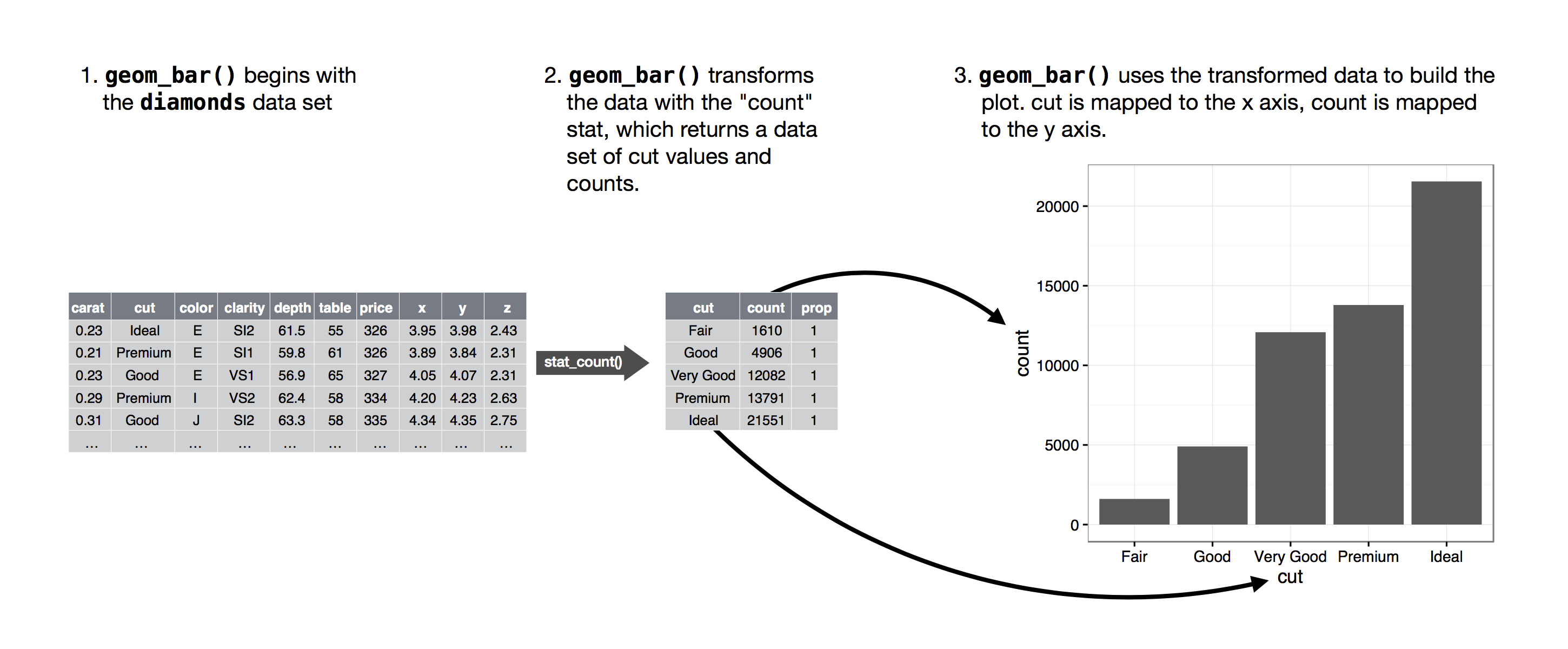
Spine charts - height version
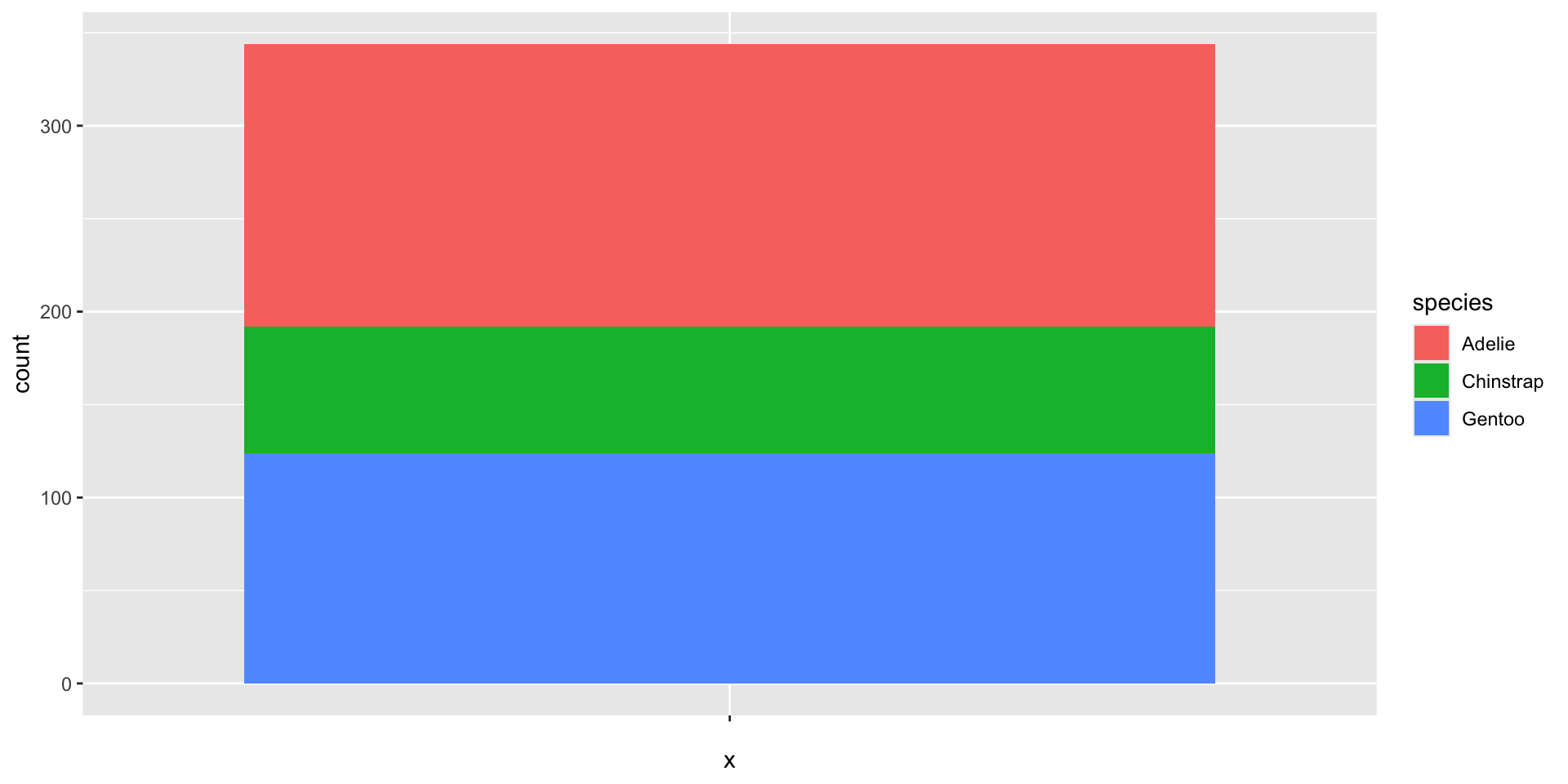
Spine charts - width version
What does a bar chart show?
Marginal Distribution
Assume categorical variable \(X\) has \(K\) categories: \(C_1, \dots, C_K\)
True marginal distribution of \(X\):
\[ P(X = C_j) = p_j,\ j \in \{ 1, \dots, K \} \]
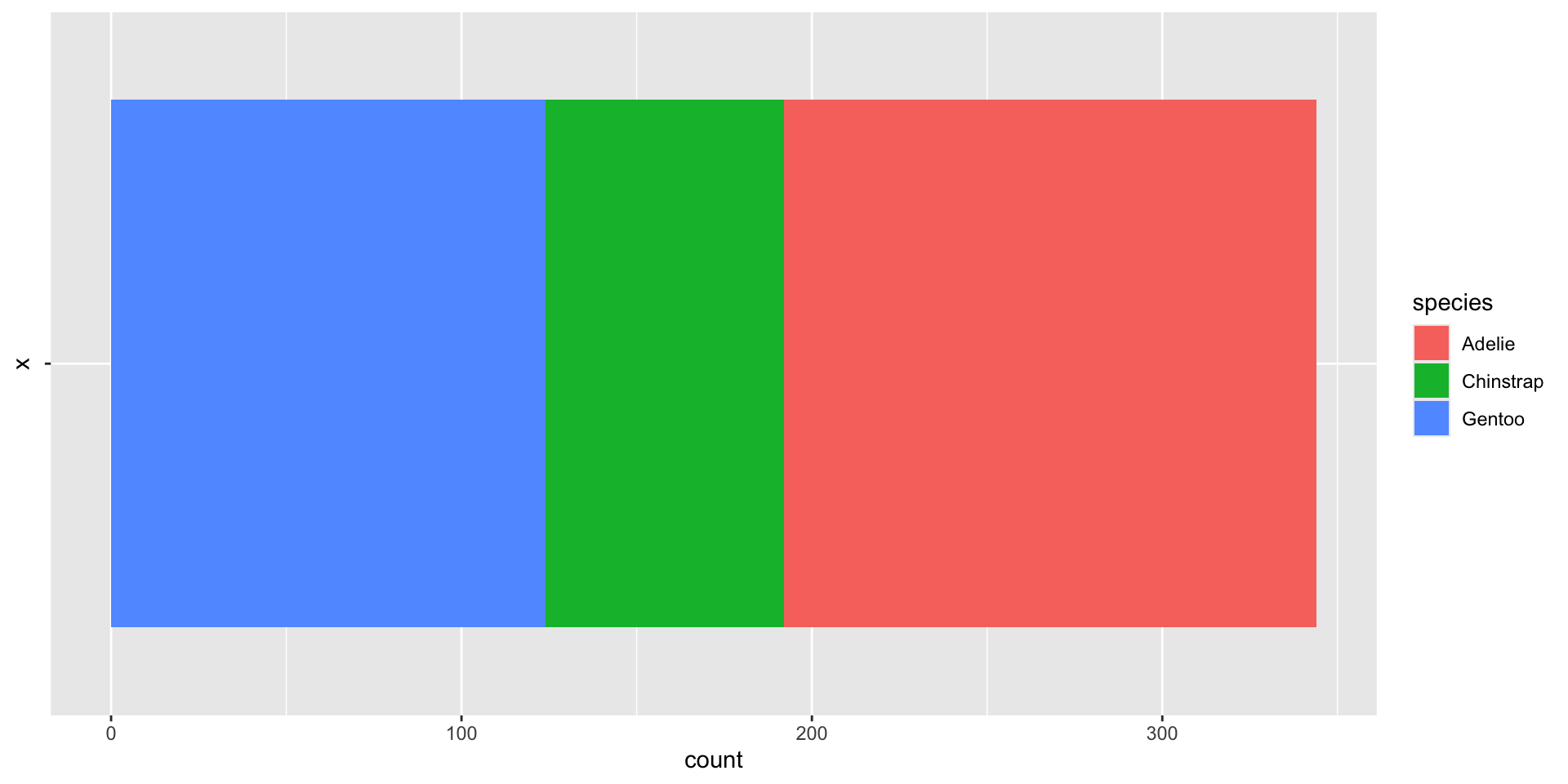
Bar charts with proportions
after_stat()indicates the aesthetic mapping is performed after statistical transformationUse
after_stat(count)to access thestat_count()called bygeom_bar()
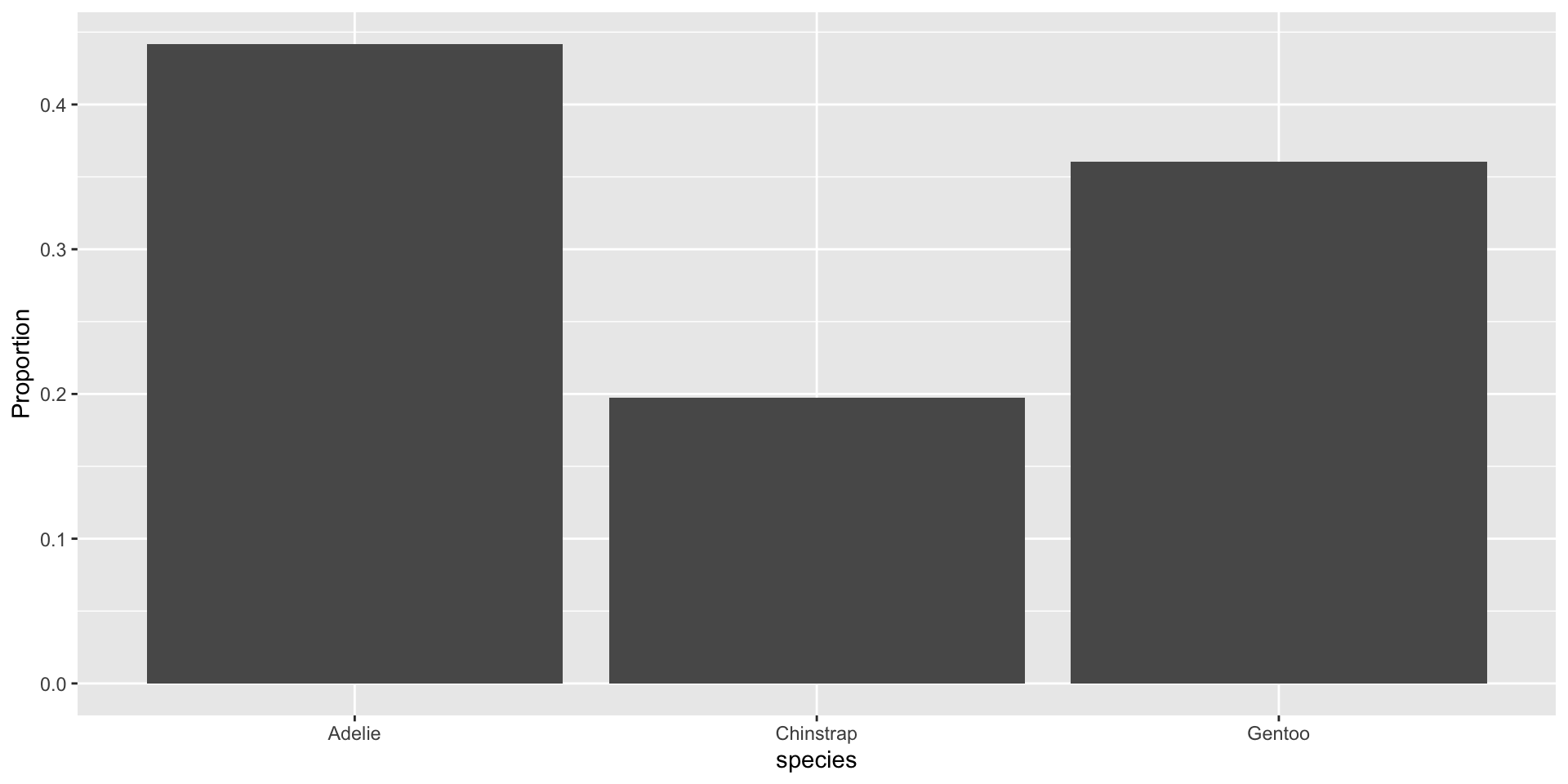
Compute and display the proportions directly
Use
group_by(),summarize(), andmutate()in a pipeline to compute then display the proportions directlyNeed to indicate we are displaying the
yaxis as given, i.e., the identity function
Compute and display the proportions directly
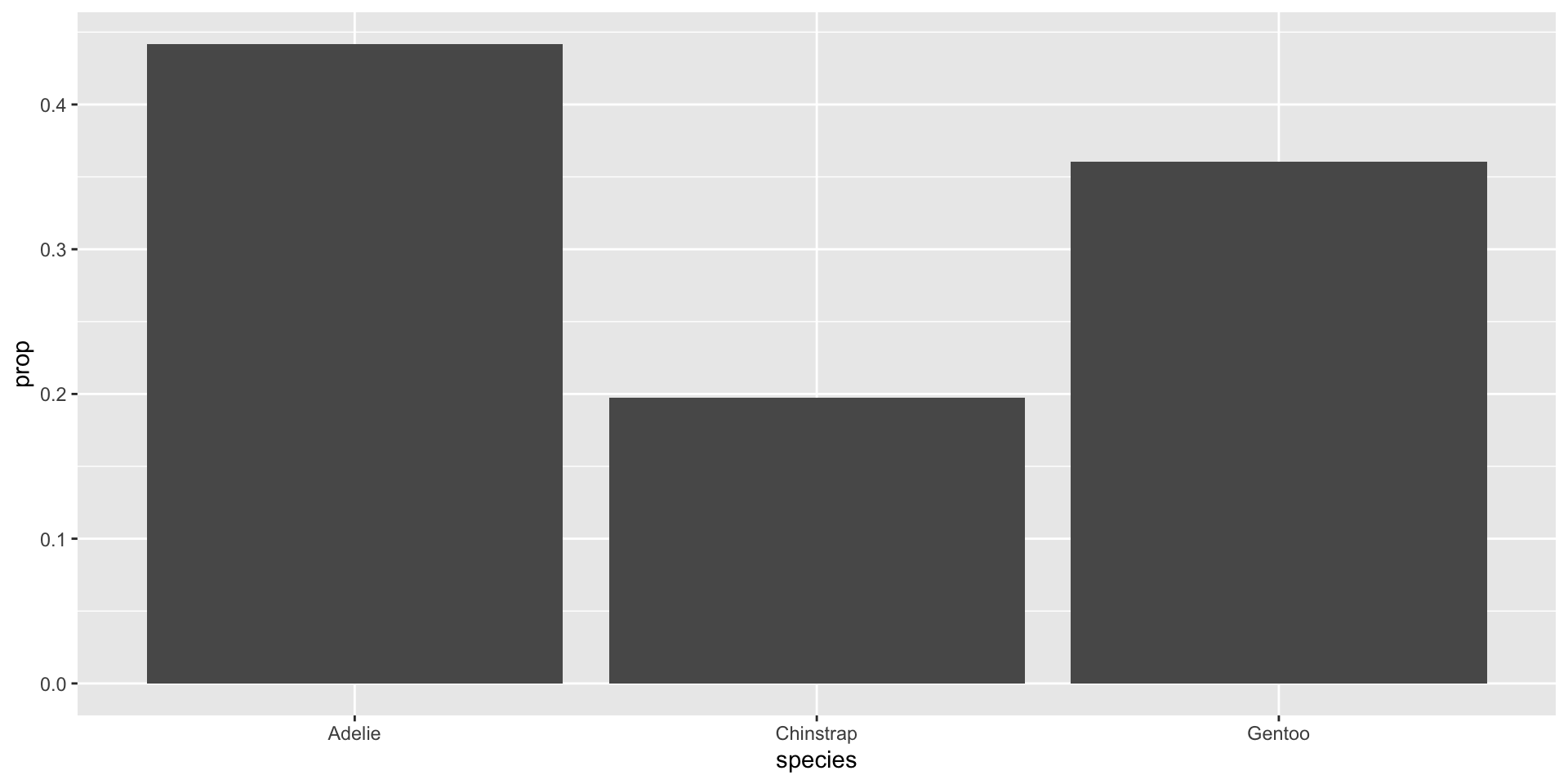
What about uncertainty?
- Quantify uncertainty for our estimate \(\hat{p}_j = \frac{n_j}{n}\) with the standard error:
\[ SE(\hat{p}_j) = \sqrt{\frac{\hat{p}_j(1 - \hat{p}_j)}{n}} \]
Compute \(\alpha\)-level confidence interval (CI) as \(\hat{p}_j \pm z_{1 - \alpha / 2} \cdot SE(\hat{p}_j)\)
Good rule-of-thumb: construct 95% CI using \(\hat{p}_j \pm 2 \cdot SE(\hat{p}_j)\)
Approximation justified by CLT, so CI could include values outside of [0,1]
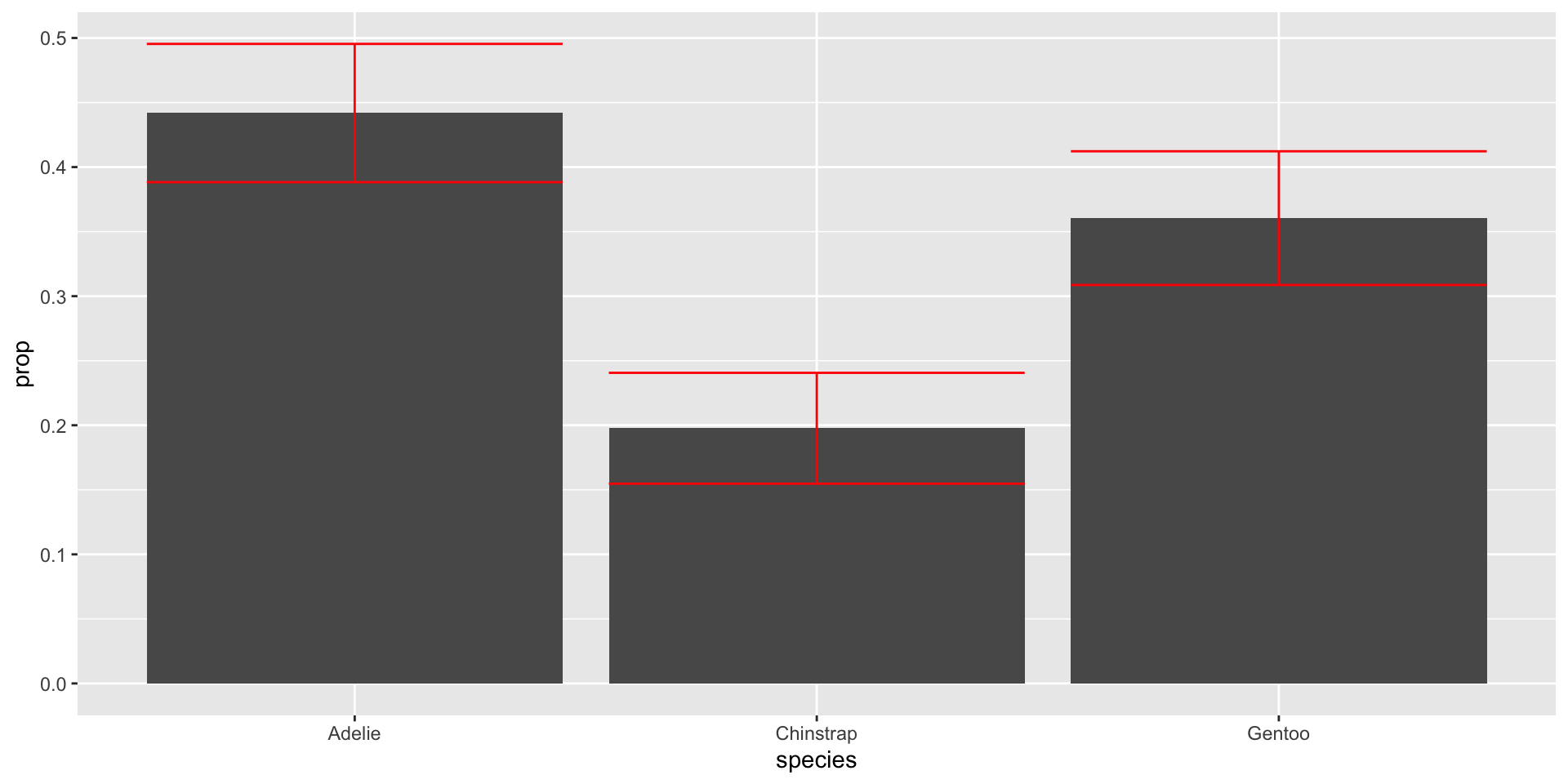
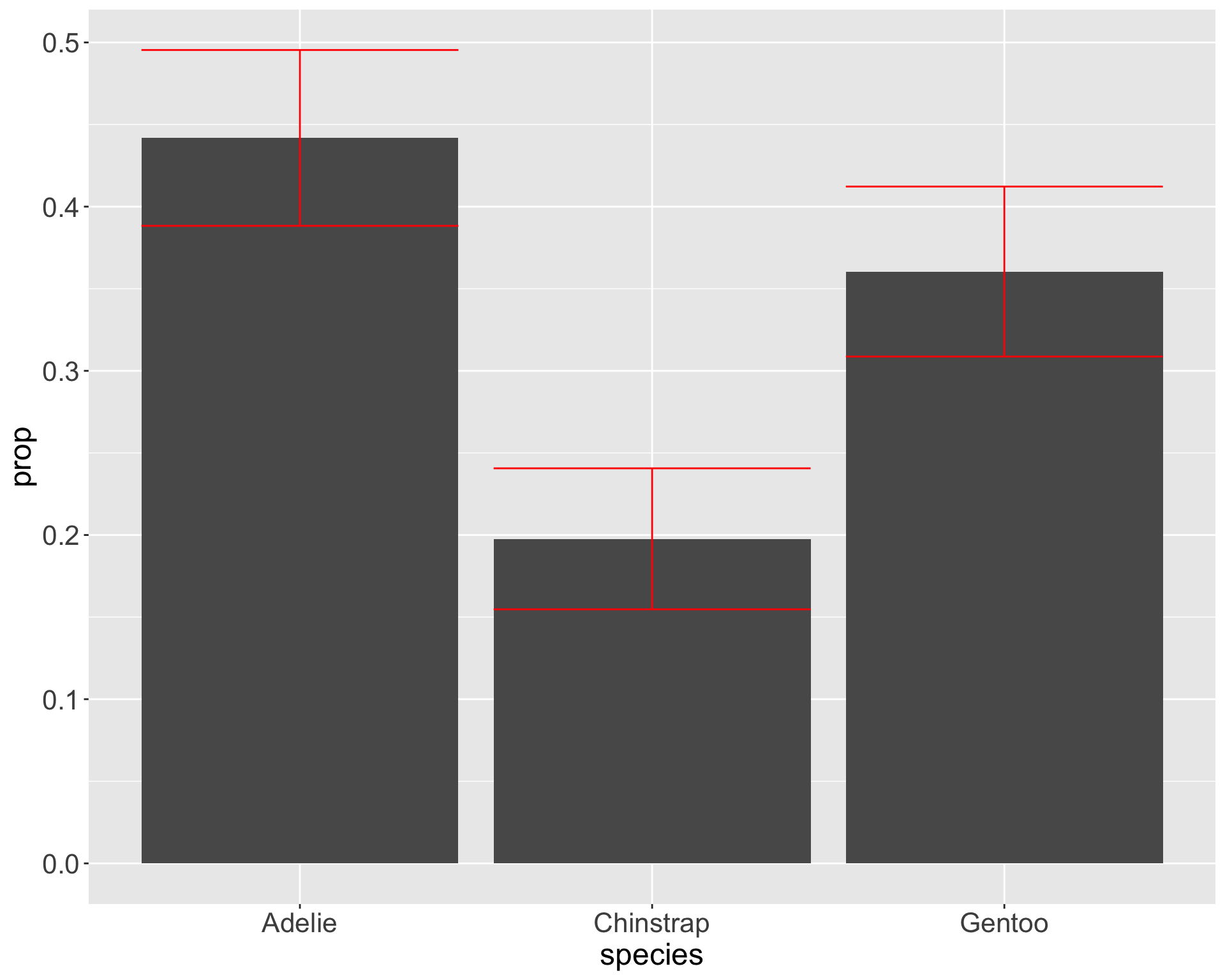
Add standard errors to bars
Need to remember each CI is for each \(\hat{p}_j\) marginally, not jointly
Have to be careful with multiple testing
penguins |>
group_by(species) |>
summarize(count = n(), .groups = "drop") |>
mutate(total = sum(count),
prop = count / total,
se = sqrt(prop * (1 - prop) / total),
lower = prop - 2 * se,
upper = prop + 2 * se) |>
ggplot(aes(x = species)) +
geom_bar(aes(y = prop), stat = "identity") +
geom_errorbar(aes(ymin = lower, ymax = upper),
color = "red") Add standard errors to bars
Why does this matter?
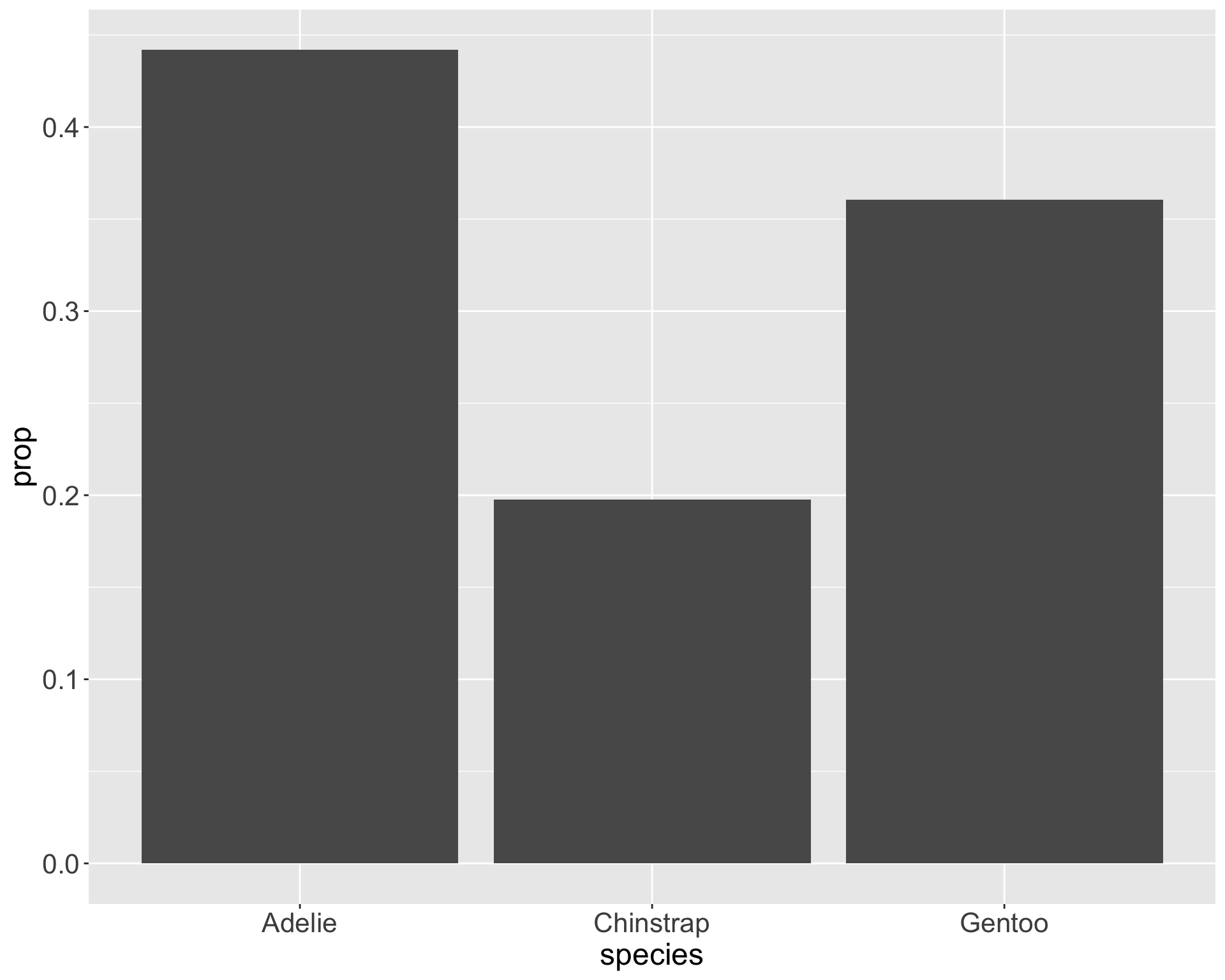
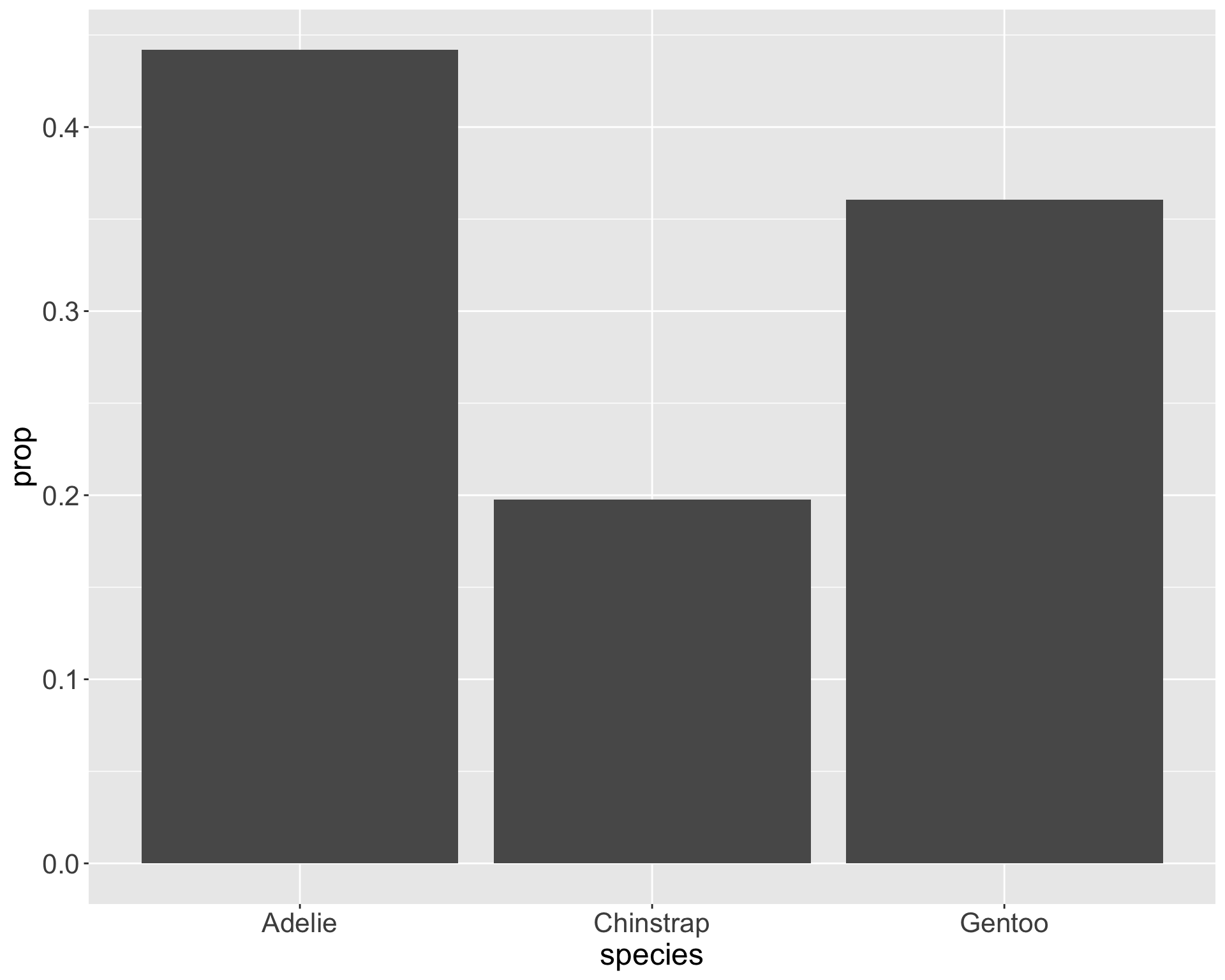
Graphs can appear the same with very different statistical conclusions - mainly due to sample size
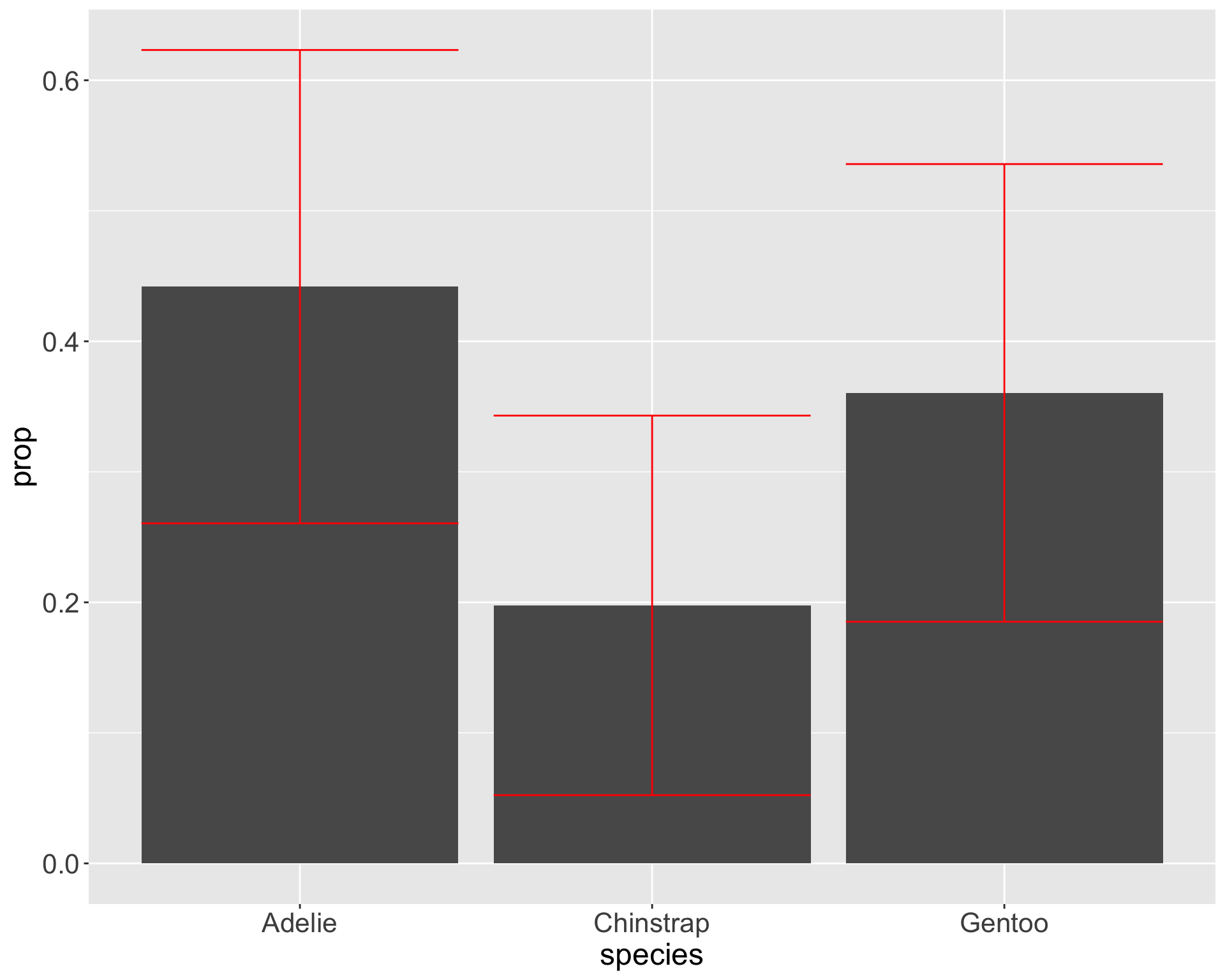
Useful to order categories by frequency with forcats
penguins |>
group_by(species) |>
summarize(count = n(), .groups = "drop") |>
mutate(total = sum(count),
prop = count / total,
se = sqrt(prop * (1 - prop) / total),
lower = prop - 2 * se,
upper = prop + 2 * se,
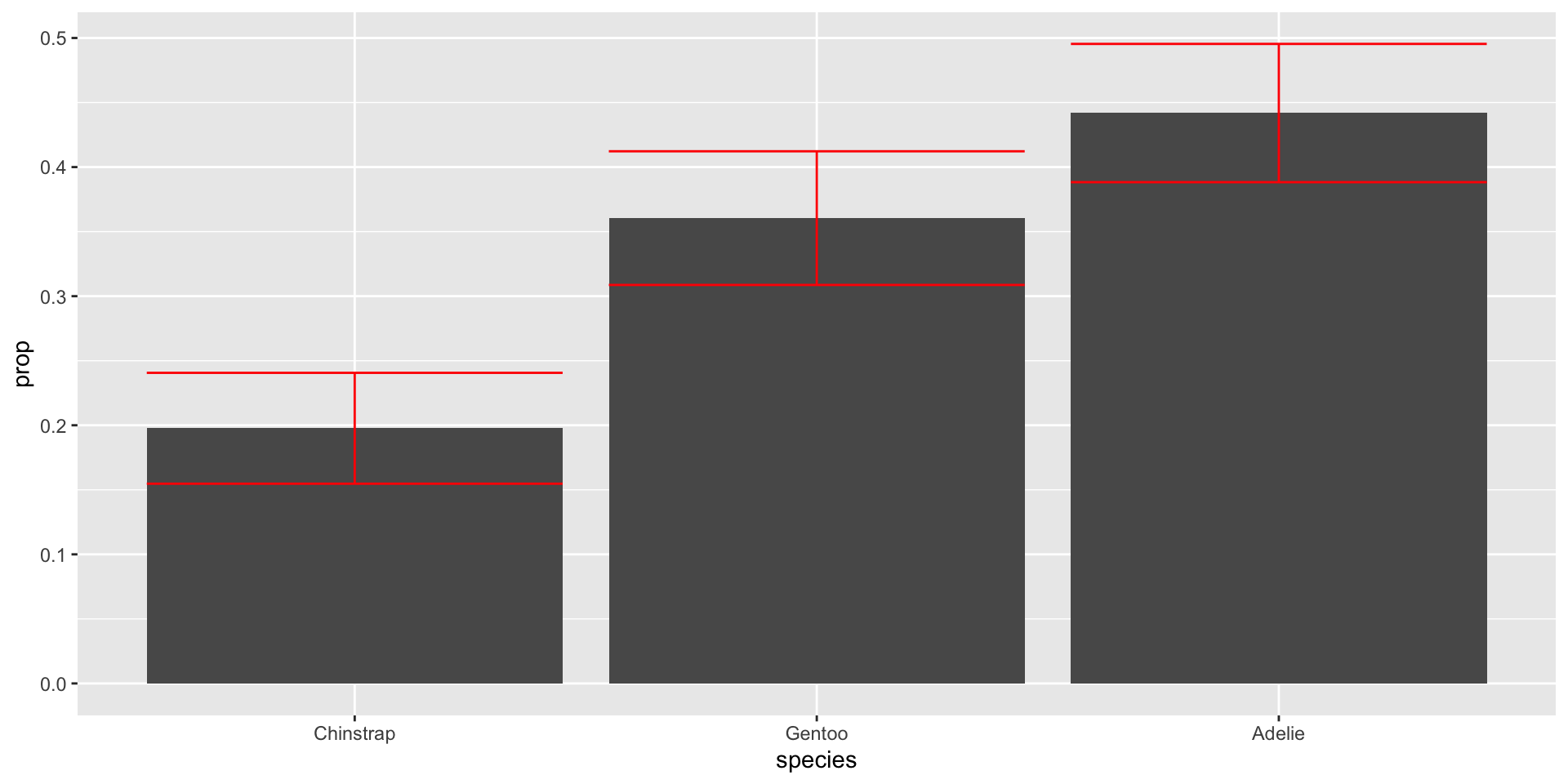
species = fct_reorder(species, prop)) |>
ggplot(aes(x = species)) +
geom_bar(aes(y = prop), stat = "identity") +
geom_errorbar(aes(ymin = lower, ymax = upper),
color = "red") Useful to order categories by frequency with forcats
So you want to make pie charts…
Friends Don’t Let Friends Make Pie Charts
Waffle charts are cooler anyway…
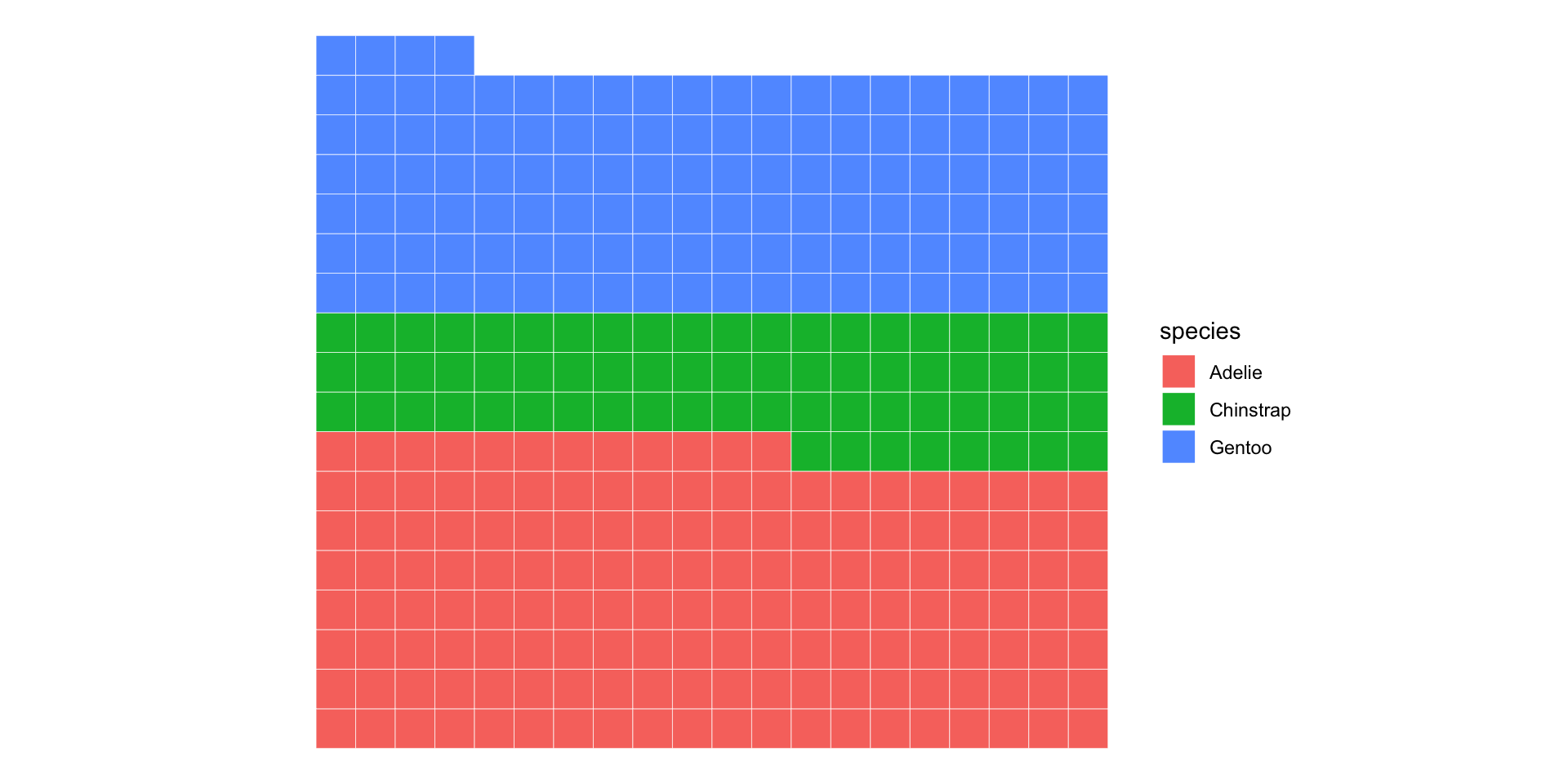
Recap and next steps
Discussed basic principles of data visualization and walked through variety of examples
Visualize categorical data with bars!
Display uncertainty with standard errors
HW1 is due next Wednesday - complete GenAI module ON TIME!
Complete HW0 by Thursday night! Confirms you have everything installed and can render
.qmdfiles to PDF viatinytex
Next time: Visualizing 2D categorical and 1D quantitative data
Recommended reading: