Visualizations for 2D Categorical and 1D Quantitative Data
2024-09-04
Reminders, previously, and today…
HW1 is due TONIGHT!
HW2 is posted (due next Wednesday)
Discussed data visualization principles and the role of infographics
Visualized 1D categorical data, i.e., make bar charts!
TODAY:
Pie charts…
Visualizing 2D categorical data
Begin 1D quantitative data
So you want to make pie charts…
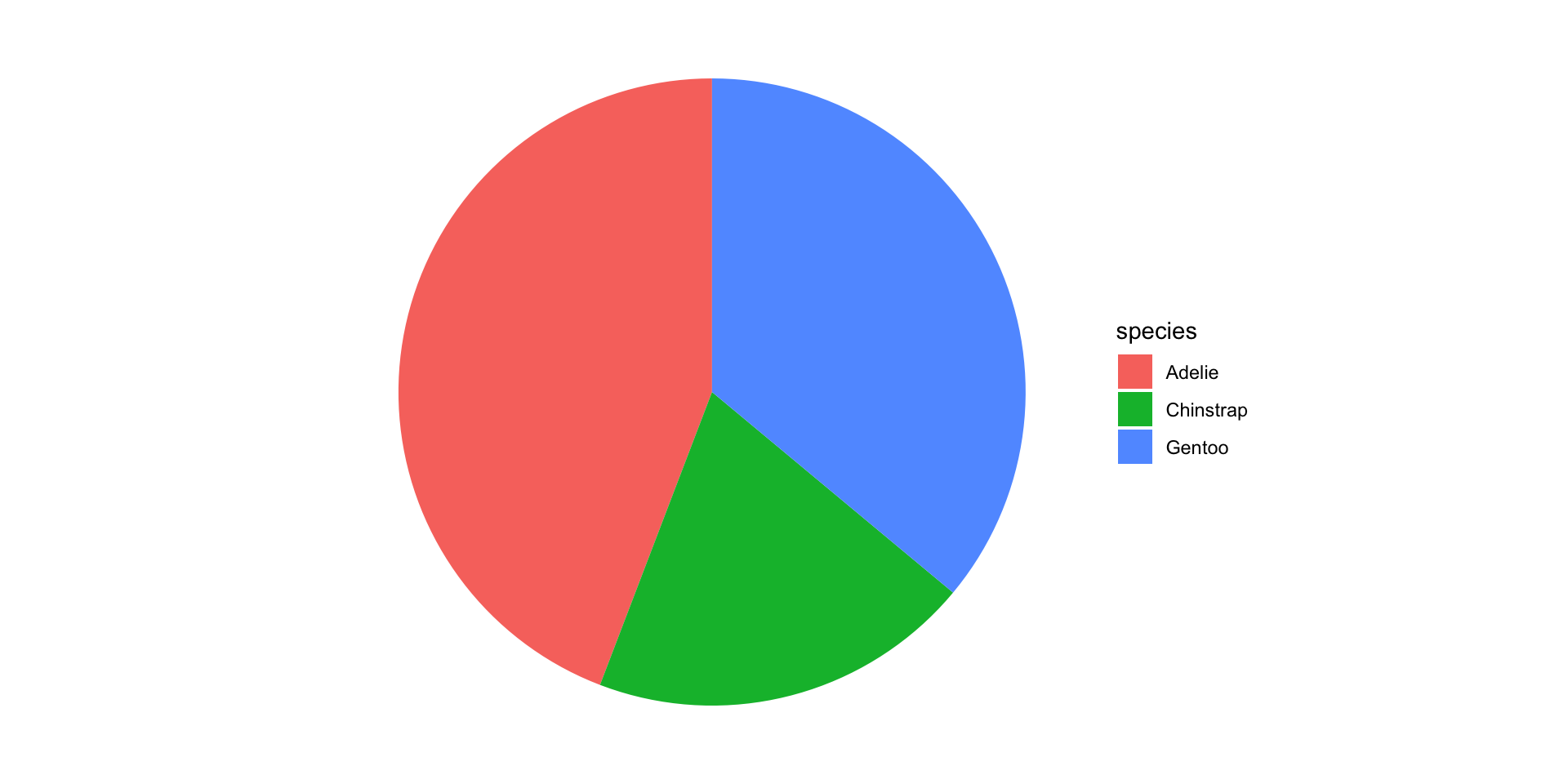
Friends Don’t Let Friends Make Pie Charts
Waffle charts are cooler anyway…
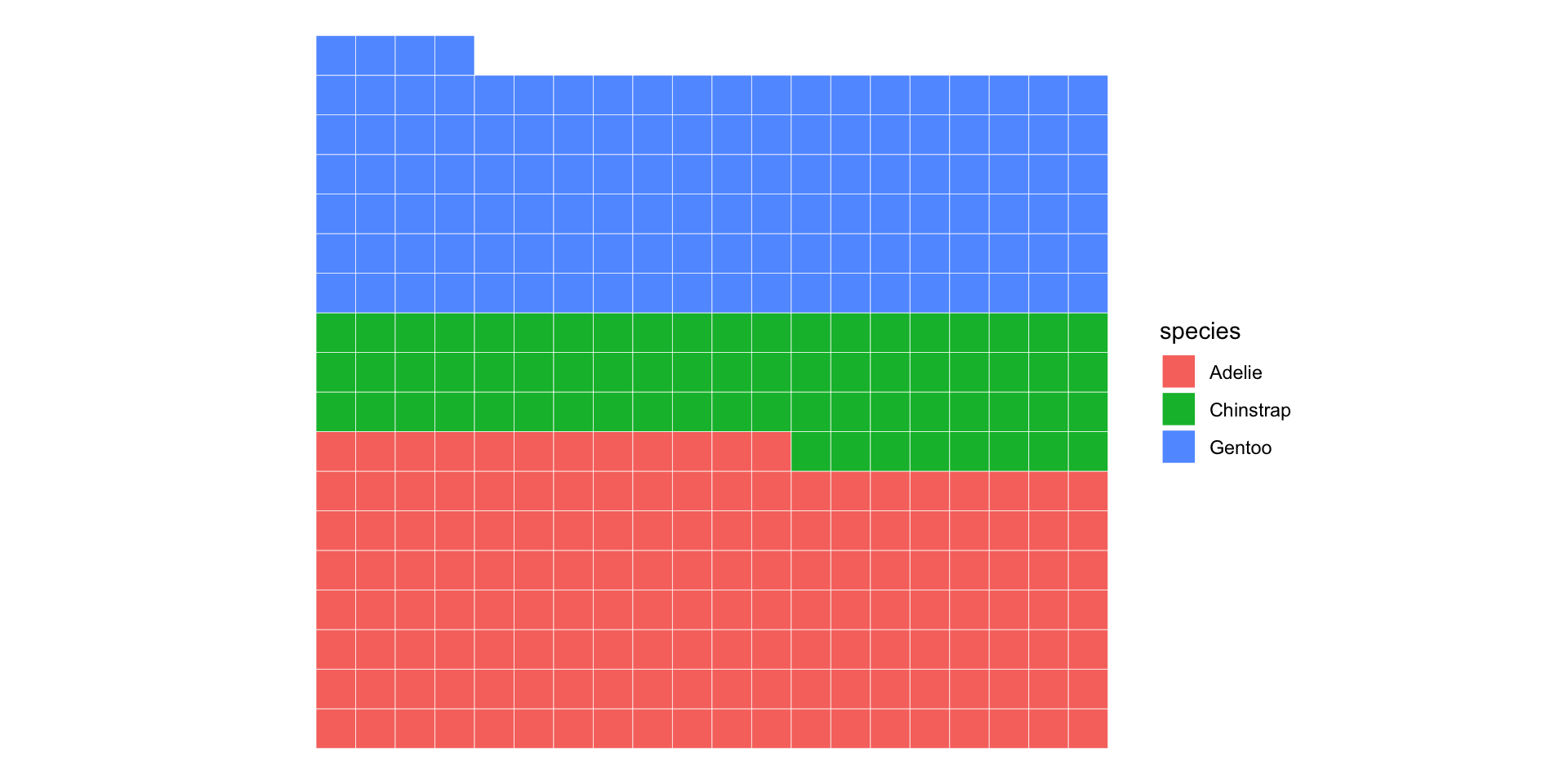
2D categorical basics: marginal / conditional distribution
Island
Species Biscoe Dream Torgersen Sum
Adelie 44 56 52 152
Chinstrap 0 68 0 68
Gentoo 124 0 0 124
Sum 168 124 52 344Column and row sums: marginal distributions
Values within rows: conditional distribution for
IslandgivenSpeciesValues within columns: conditional distribution for
SpeciesgivenIslandBottom right: total number of observations
Connecting distributions to visualizations
Five distributions for two categorical variables \(A\) and \(B\):
Marginals: \(P(A)\) and \(P(B)\)
Conditionals: \(P(A | B)\) and \(P(B|A)\)
Joint: \(P(A, B)\)
We use bar charts to visualize marginal distributions for categorical variables…
And we’ll use more bar charts to visualize conditional and joint distributions!
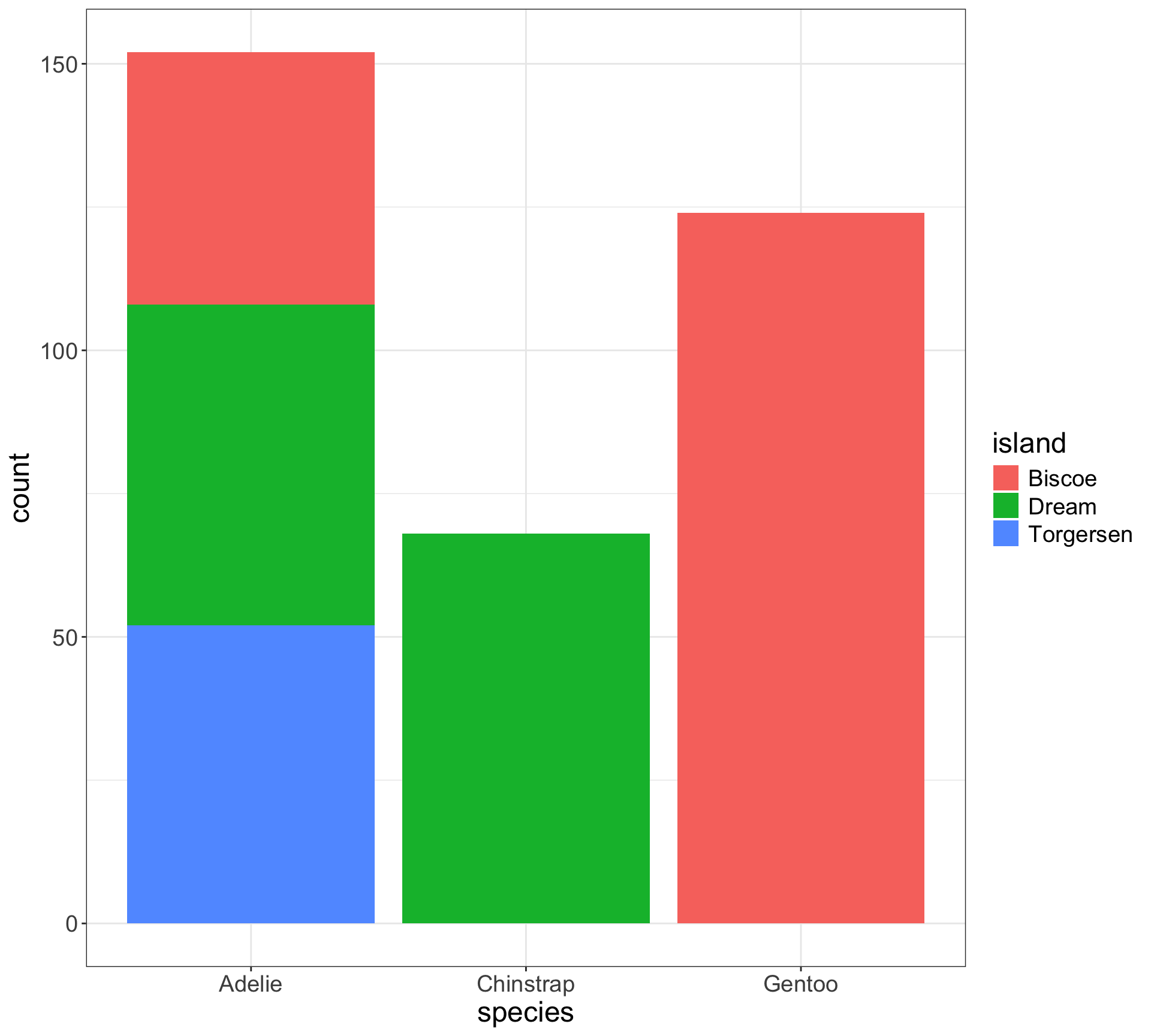
Stacked bar charts - a bar chart of spine charts
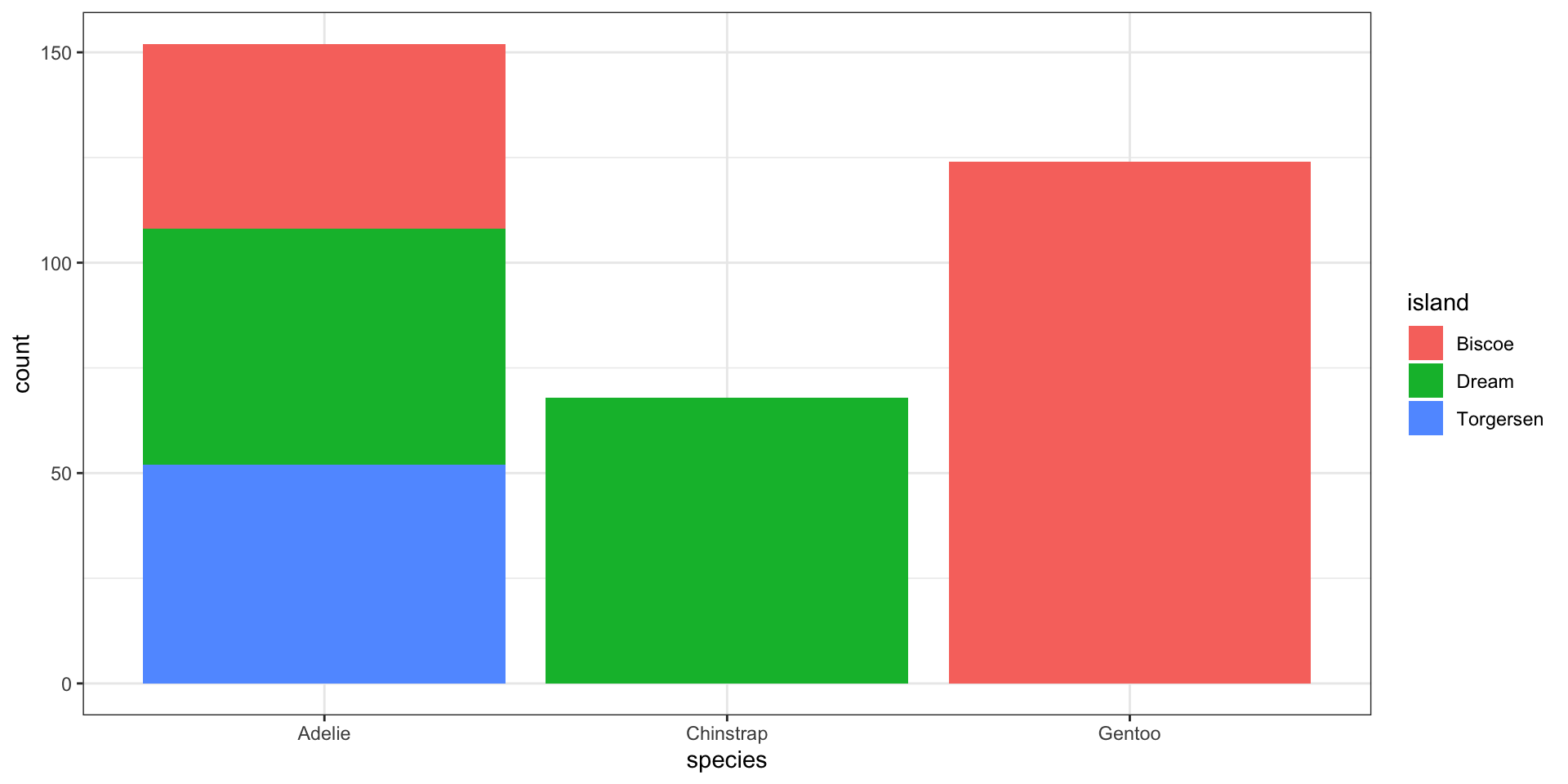
Easy to see marginal of
species, i.e., \(P(\)x\()\)Can see conditional of
island|species, i.e., \(P(\)fill|x\()\)Harder to see conditional of
species|island, i.e., \(P(\)x|fill\()\)
Side-by-side bar charts
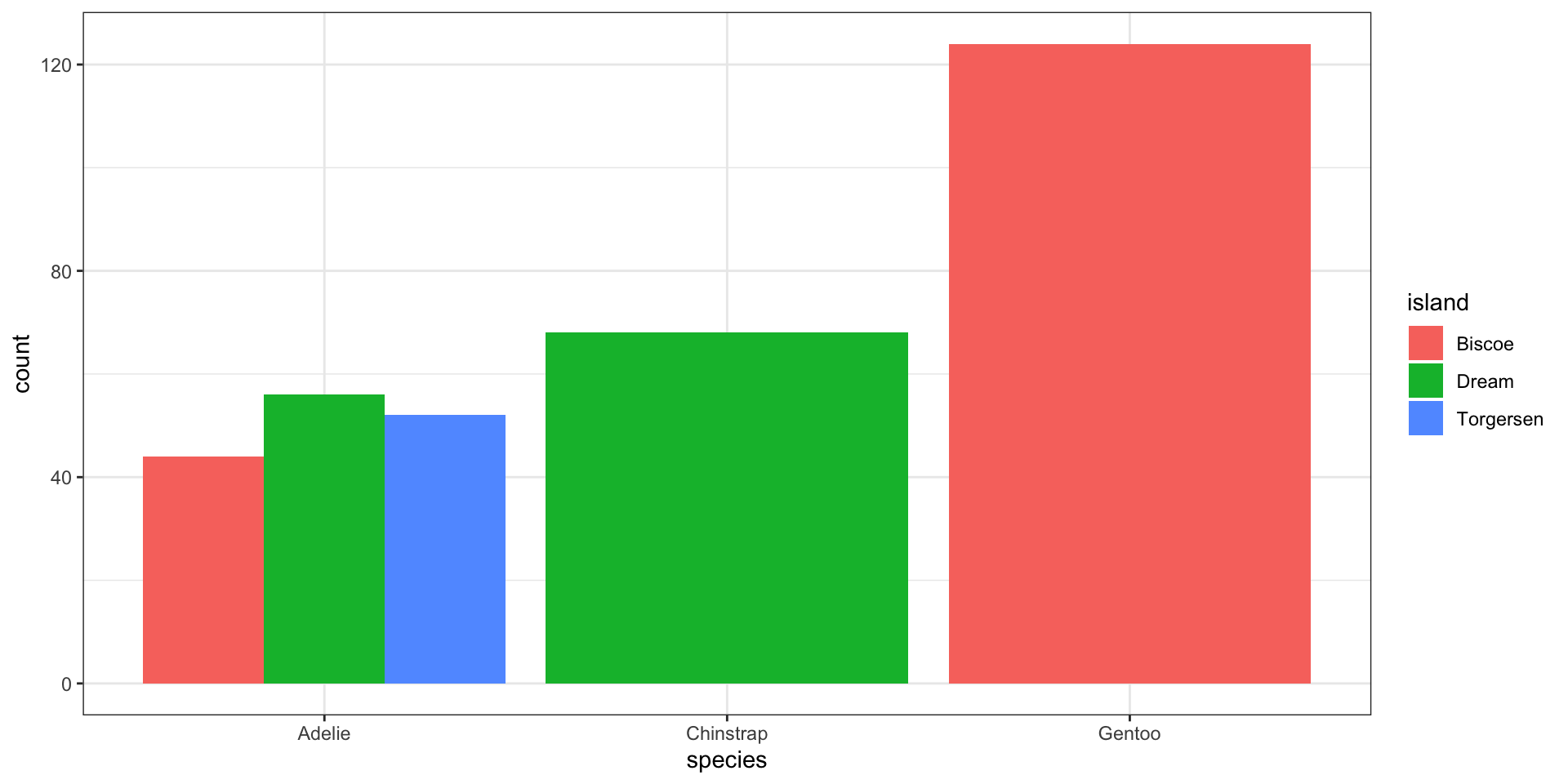
Easy to see conditional of
island|species, i.e., \(P(\)fill|x\()\)Can see conditional of
species|island, i.e., \(P(\)x|fill\()\)
Side-by-side bar charts
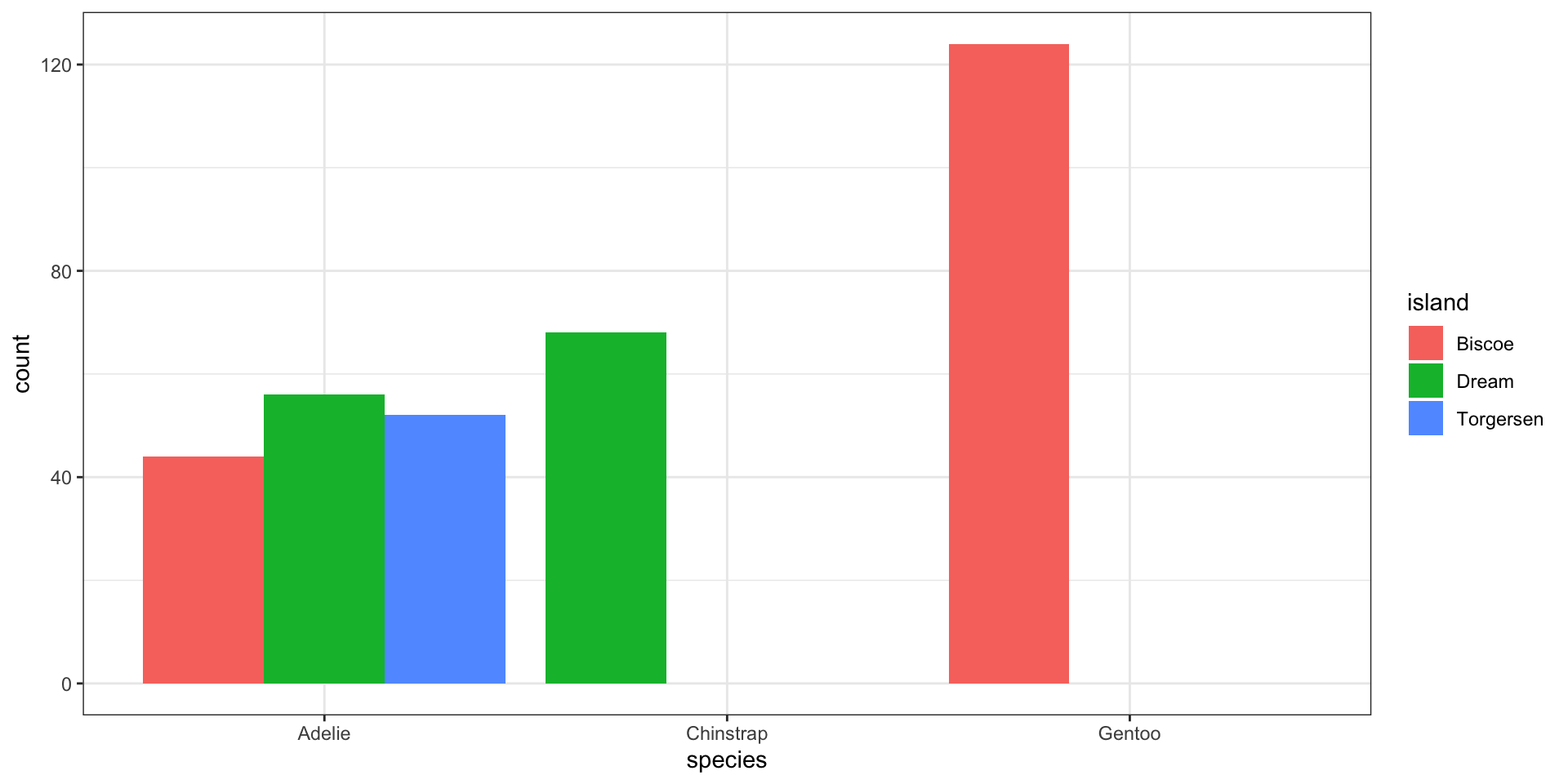
Easy to see conditional of
island|species, i.e., \(P(\)fill|x\()\)Can see conditional of
species|island, i.e., \(P(\)x|fill\()\)
Complete missing values to preserve location
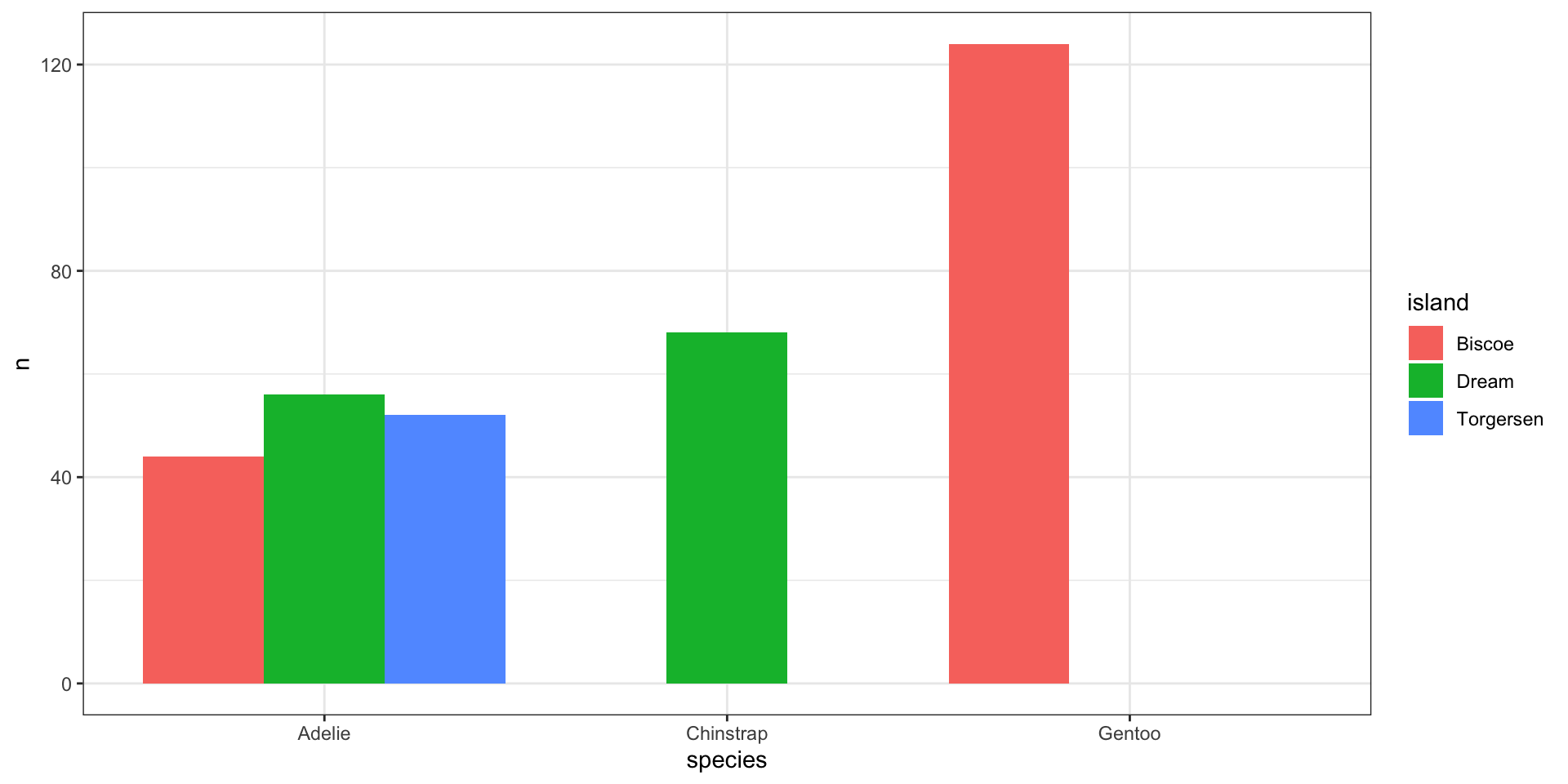
What do you prefer?
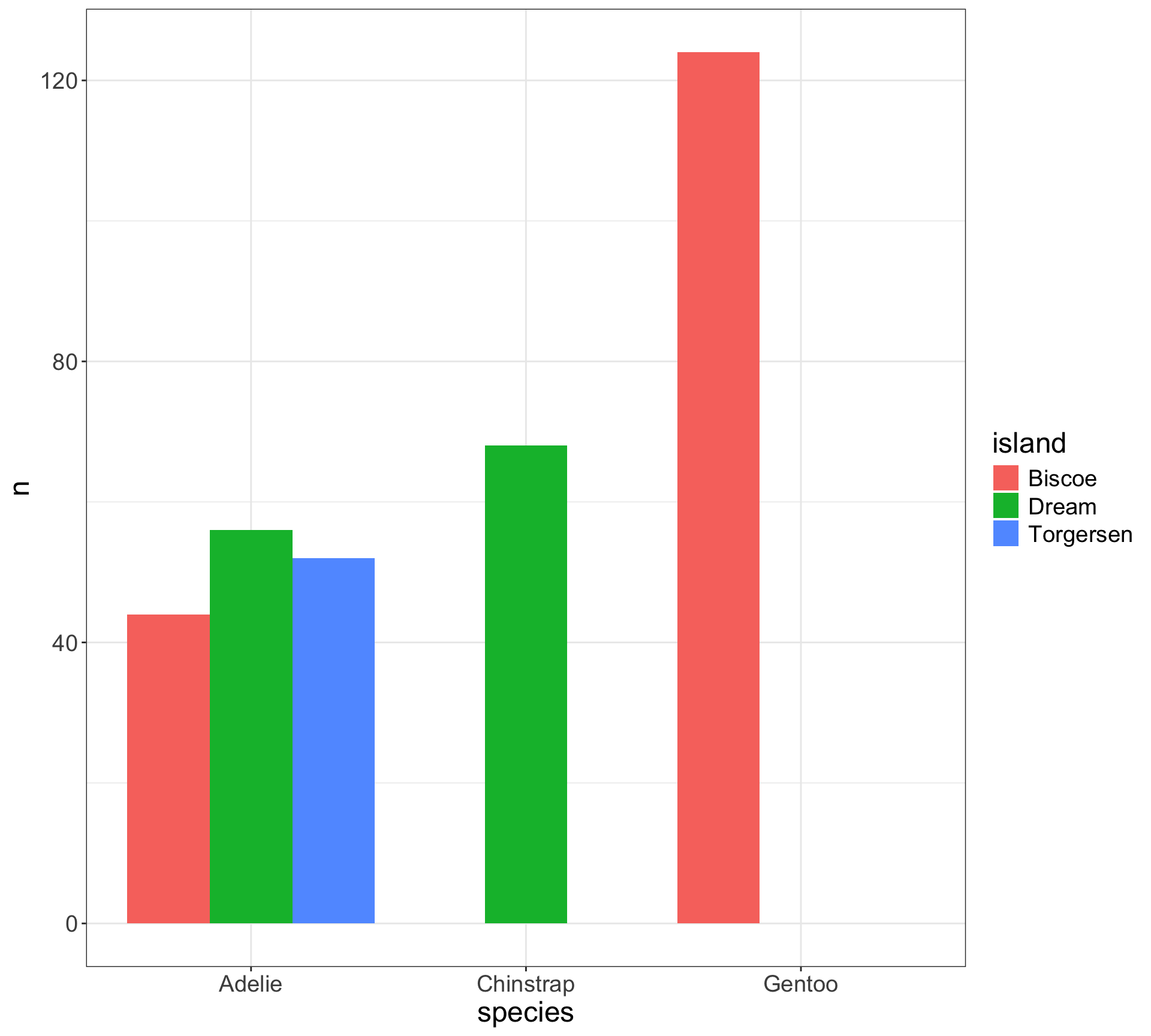
Chi-squared test for 1D categorical data:
- Null hypothesis \(H_0\): \(p_1 = p_2 = \dots = p_K\), compute the test statistic:
\[ \chi^2 = \sum_{j=1}^K \frac{(O_j - E_j)^2}{E_j} \]
\(O_j\): observed counts in category \(j\)
\(E_j\): expected counts under \(H_0\), i.e., each category is equally to occur \(n / K = p_1 = p_2 = \dots = p_K\)
Hypothesis testing review
Computing \(p\)-values works like this:
Choose a test statistic.
Compute the test statistic in your dataset.
Is test statistic “unusual” compared to what I would expect under \(H_0\)?
Compare \(p\)-value to target error rate \(\alpha\) (typically referred to as target level \(\alpha\) )
Typically choose \(\alpha = 0.05\)
- i.e., if we reject null hypothesis at \(\alpha = 0.05\) then, assuming \(H_0\) is true, there is a 5% chance it is a false positive (aka Type 1 error)

Inference for 2D categorical data
Again we use the chi-squared test:
- Null hypothesis \(H_0\): variables \(A\) and \(B\) are independent, compute the test statistic:
\[\chi^2 = \sum_{i}^{K_A} \sum_{j}^{K_B} \frac{(O_{ij} - E_{ij})^2}{E_{ij}}\]
\(O_{ij}\): observed counts in contingency table
\(E_{ij}\): expected counts under \(H_0\)
\[ \begin{aligned} E_{ij} &= n \cdot P(A = a_i, B = b_j) \\ &= n \cdot P(A = a_i) P(B = b_j) \\ &= n \cdot \left( \frac{n_{i \cdot}}{n} \right) \left( \frac{ n_{\cdot j}}{n} \right) \end{aligned} \]
Inference for 2D categorical data
Again we use the chi-squared test:
- Null hypothesis \(H_0\): variables \(A\) and \(B\) are independent, compute the test statistic:
\[\chi^2 = \sum_{i}^{K_A} \sum_{j}^{K_B} \frac{(O_{ij} - E_{ij})^2}{E_{ij}}\]
\(O_{ij}\): observed counts in contingency table
\(E_{ij}\): expected counts under \(H_0\)
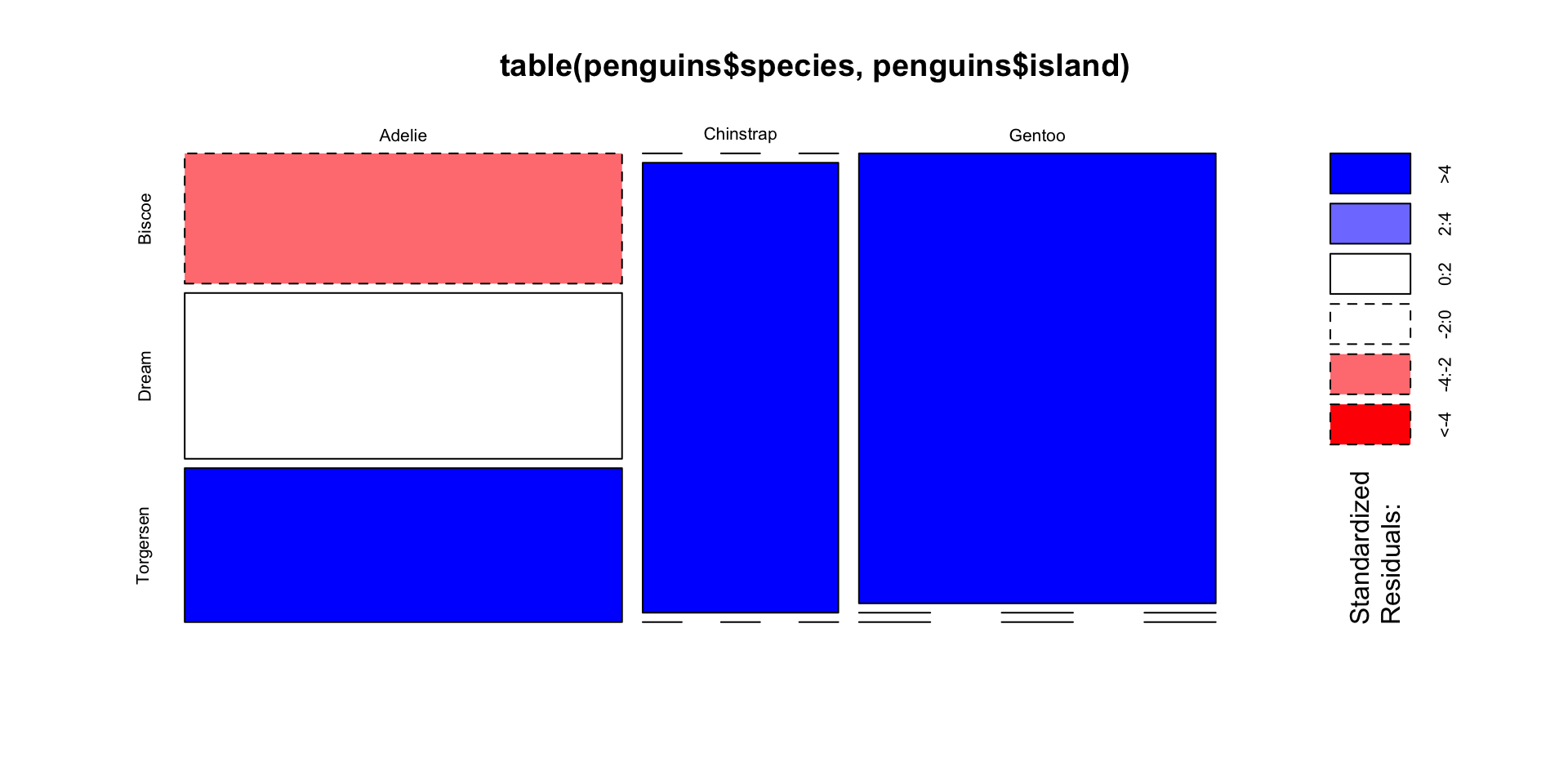
Visualize independence test with mosaic plots
Two variables are independent if knowing the level of one tells us nothing about the other
- i.e. \(P(A | B) = P(A)\), and that \(P(A, B) = P(A) \times P(B)\)
Create a mosaic plot using base R

Shade by Pearson residuals
- The test statistic is:
\[\chi^2 = \sum_{i}^{K_A} \sum_{j}^{K_B} \frac{(O_{ij} - E_{ij})^2}{E_{ij}}\]
- Define the Pearson residuals as:
\[r_{ij} = \frac{O_{ij} - E_{ij}}{\sqrt{E_{ij}}}\]
- Side-note: In general, Pearson residuals are \(\frac{\text{residuals}}{\sqrt{\text{variance}}}\)
\(r_{ij} \approx 0 \rightarrow\) observed counts are close to expected counts
\(|r_{ij}| > 2 \rightarrow\) “significant” at level \(\alpha = 0.05\).
Very positive \(r_{ij} \rightarrow\) more than expected, while very negative \(r_{ij} \rightarrow\) fewer than expected
Color by Pearson residuals to tell us which combos are much bigger/smaller than expected.
Bonus: Treemaps do not require same categorical levels across subgroups
library(treemapify)
penguins |>
group_by(species, island) |>
summarize(count = n(), .groups = "drop") |>
ggplot(aes(area = count, subgroup = island,
label = species,
fill = interaction(species, island))) +
# 1. Draw species borders and fill colors
geom_treemap() +
# 2. Draw island borders
geom_treemap_subgroup_border() +
# 3. Print island text
geom_treemap_subgroup_text(place = "centre", grow = T,
alpha = 0.5, colour = "black",
fontface = "italic", min.size = 0) +
# 4. Print species text
geom_treemap_text(colour = "white", place = "topleft",
reflow = T) +
guides(colour = "none", fill = "none")Bonus: Treemaps do not require same categorical levels across subgroups

1D Quantitative Data
Observations are collected into a vector \((x_1, \dots, x_n)\), \(x_i \in \mathbb{R}\) (or \(\mathbb{R}^+\), \(\mathbb{Z}\))
Common summary statistics for 1D quantitative data:
Center: Mean, median, weighted mean, mode
- Related to the first moment, i.e., \(\mathbb{E}[X]\)
Spread: Variance, range, min/max, quantiles, IQR
- Related to the second moment, i.e., \(\mathbb{E}[X^2]\)
Shape: symmetry, skew, kurtosis (“peakedness”)
- Related to higher order moments, i.e., skewness is \(\mathbb{E}[X^3]\), kurtosis is \(\mathbb{E}[X^4]\)
Compute various statistics with summary(), mean(), median(), quantile(), range(), sd(), var(), etc.
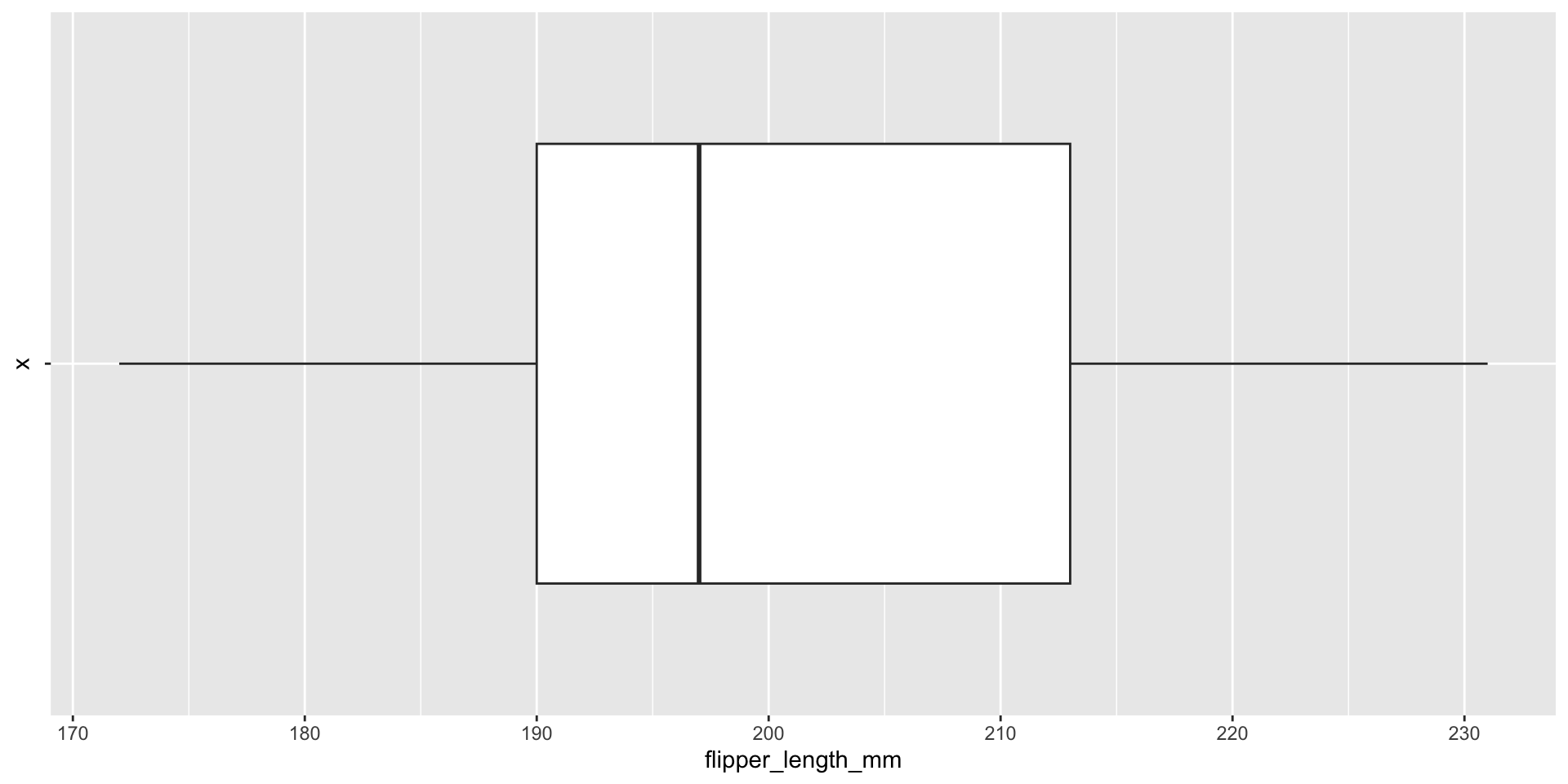
Box plots visualize summary statistics
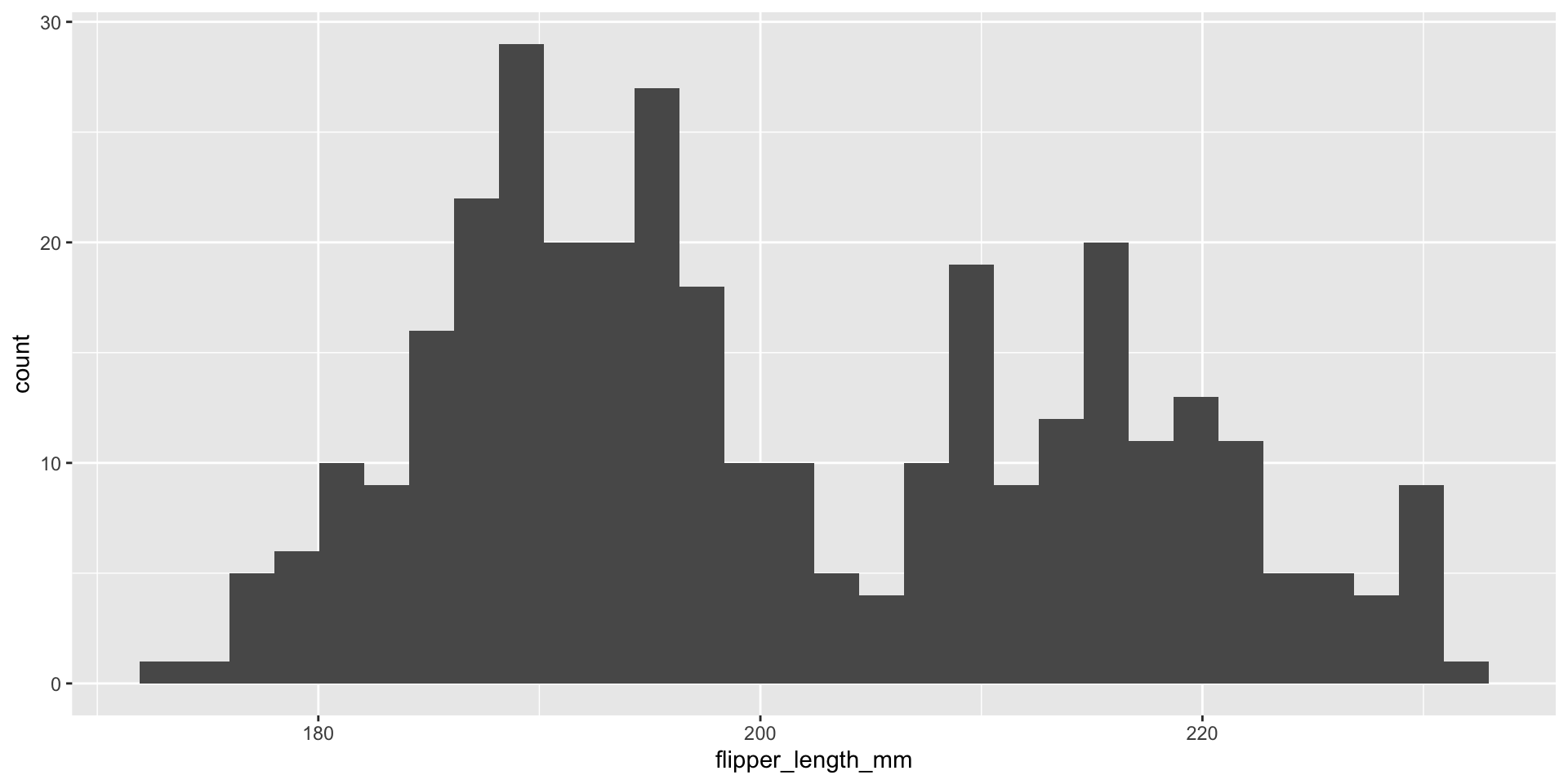
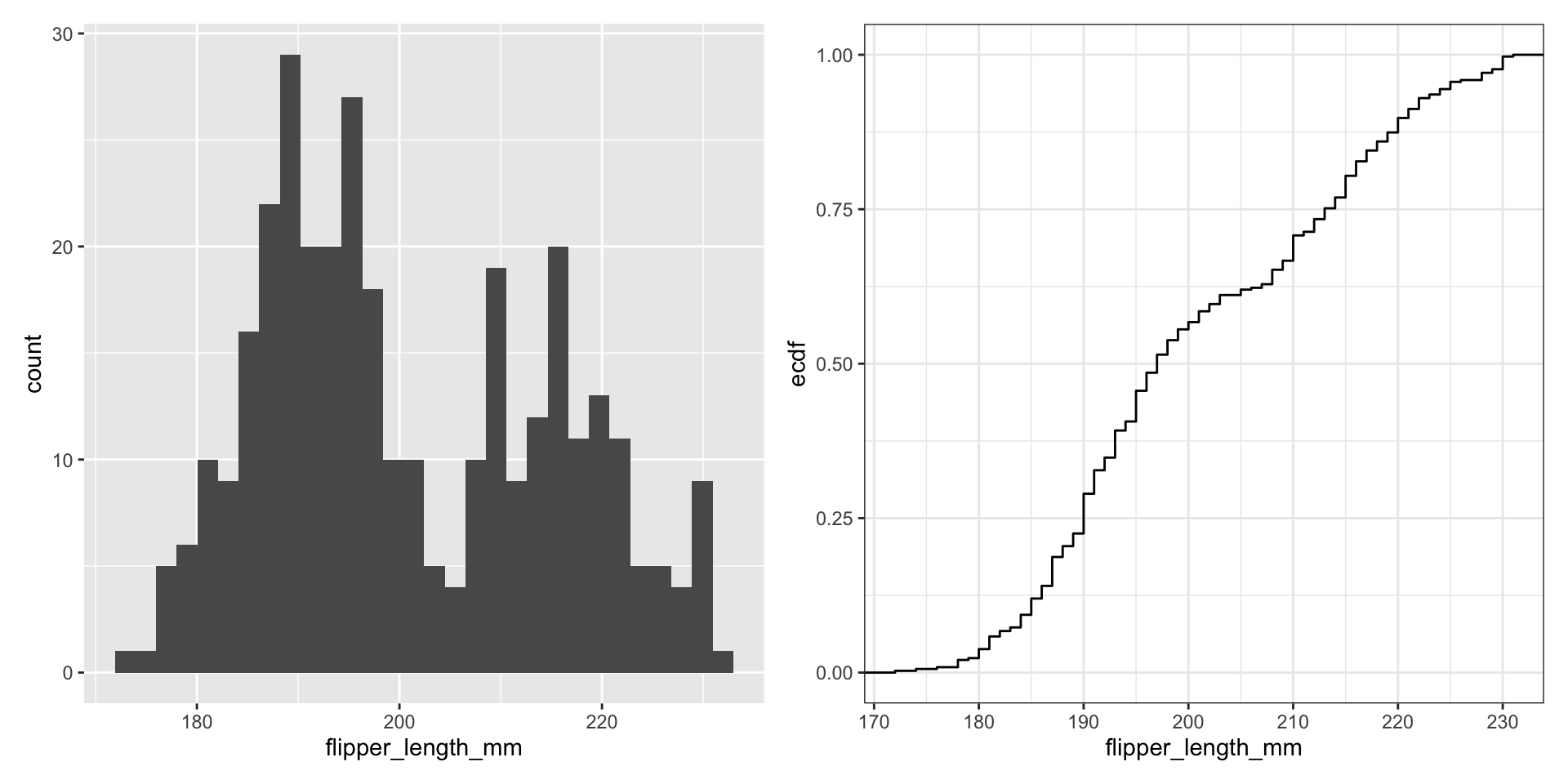
Histograms display 1D continuous distributions
Do NOT rely on box plots…
What do visualizations of continuous distributions display?
Probability that continuous variable \(X\) takes a particular value is 0
e.g., \(P\) (flipper_length_mm \(= 200\)) \(= 0\), why?
Instead we use the probability density function (PDF) to provide a relative likelihood
For continuous variables we can use the cumulative distribution function (CDF),
\[ F(x) = P(X \leq x) \]
For \(n\) observations we can easily compute the Empirical CDF (ECDF):
\[\hat{F}_n(x) = \frac{\text{# obs. with variable} \leq x}{n} = \frac{1}{n} \sum_{i=1}^{n}1(x_i \leq x)\]
- where \(1()\) is the indicator function, i.e.
ifelse(x_i <= x, 1, 0)
Display full distribution with ECDF plot
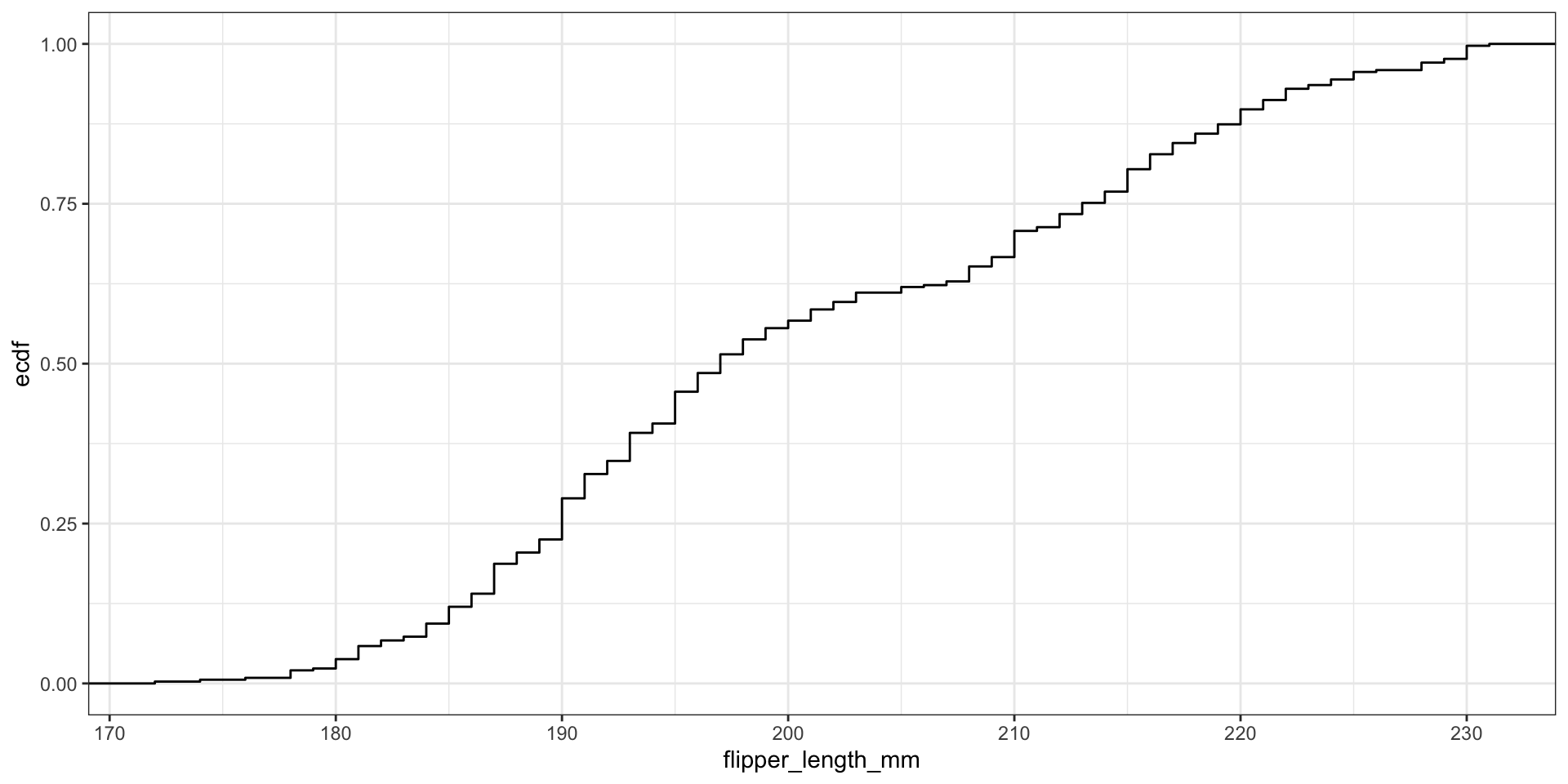
What’s the relationship between these two figures?
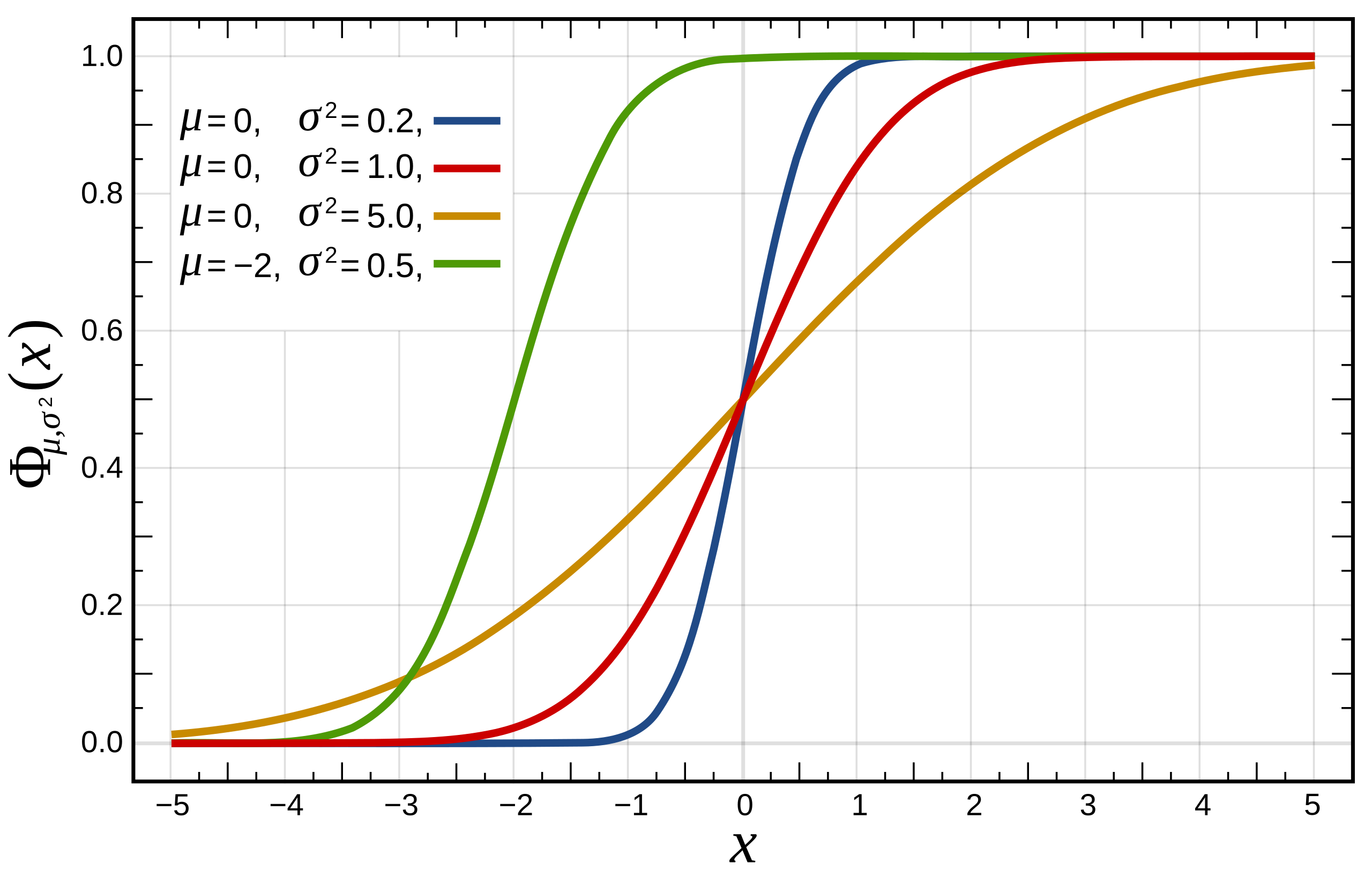
What about comparing to theoretical distributions?

One-Sample Kolmogorov-Smirnov Test
We compare the ECDF \(\hat{F}(x)\) to a theoretical distribution’s CDF \(F(x)\)
The one sample KS test statistic is: \(\text{max}_x |\hat{F}(x) - F(x)|\)

Flipper length example
What if we assume flipper_length_mm follows Normal distribution?
- i.e.,
flipper_length_mm\(\sim N(\mu, \sigma^2)\)
Need estimates for mean \(\mu\) and standard deviation \(\sigma\):
Perform one-sample KS test using ks.test():
Flipper length example
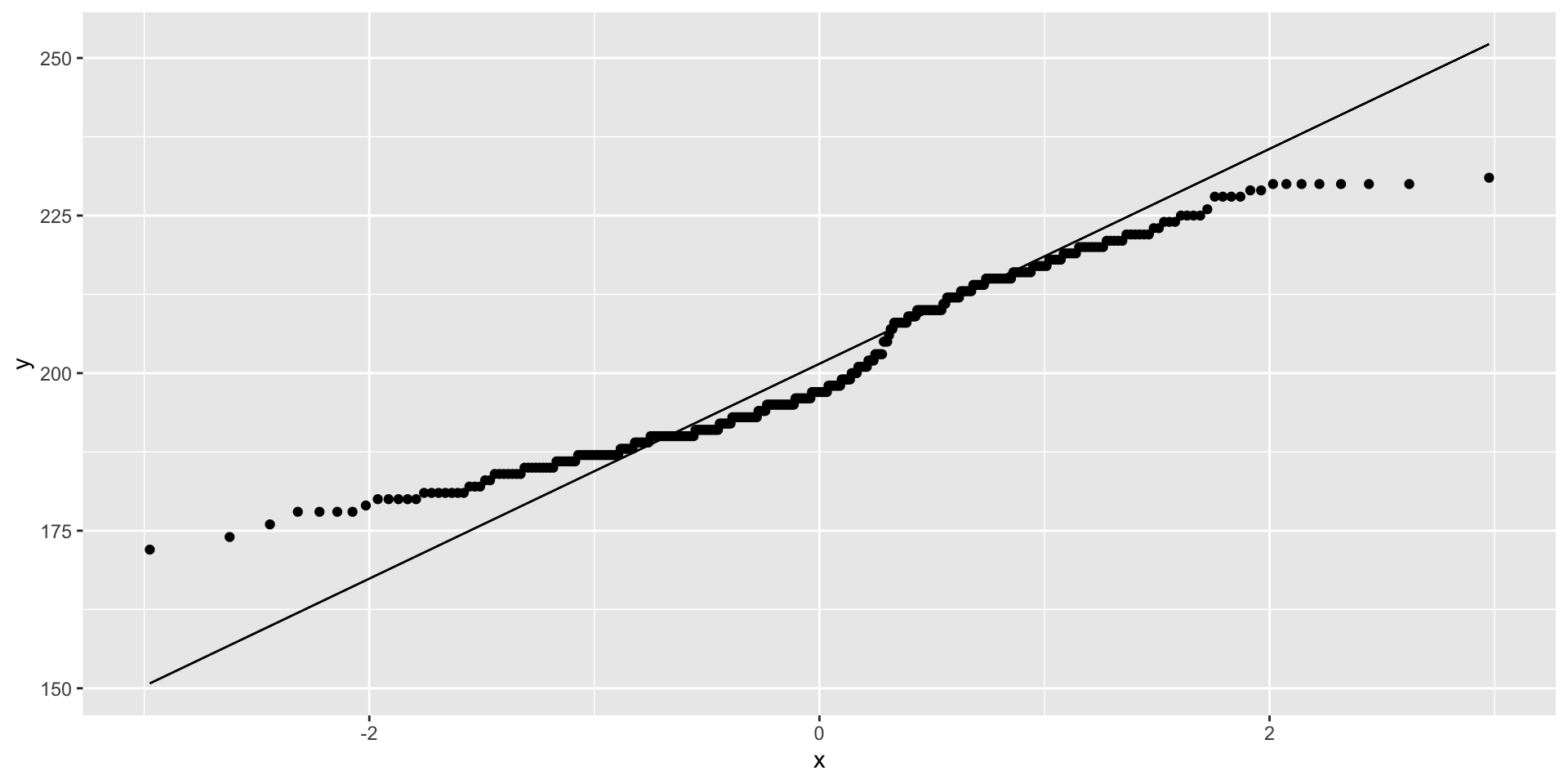
Visualize distribution comparisons using quantile-quantile (q-q) plots
Recap and next steps
Visualize categorical data with bars! Regular, stacked, side-by-side, mosaic
Display uncertainty: (1D) standard errors, (2D) Pearson residuals
Visualize 1D quantitative data with histograms, ECDFs, but never use a box plot by itself
HW1 is due TONIGHT!
HW2 is posted (due next Wednesday)
Next time: Density Estimation
Recommended reading:
CW Chapter 11 Visualizing nested proportions, CW Chapter 7 Visualizing distributions: Histograms and density plots, CW Chapter 8 Visualizing distributions: Empirical cumulative distribution functions and q-q plots
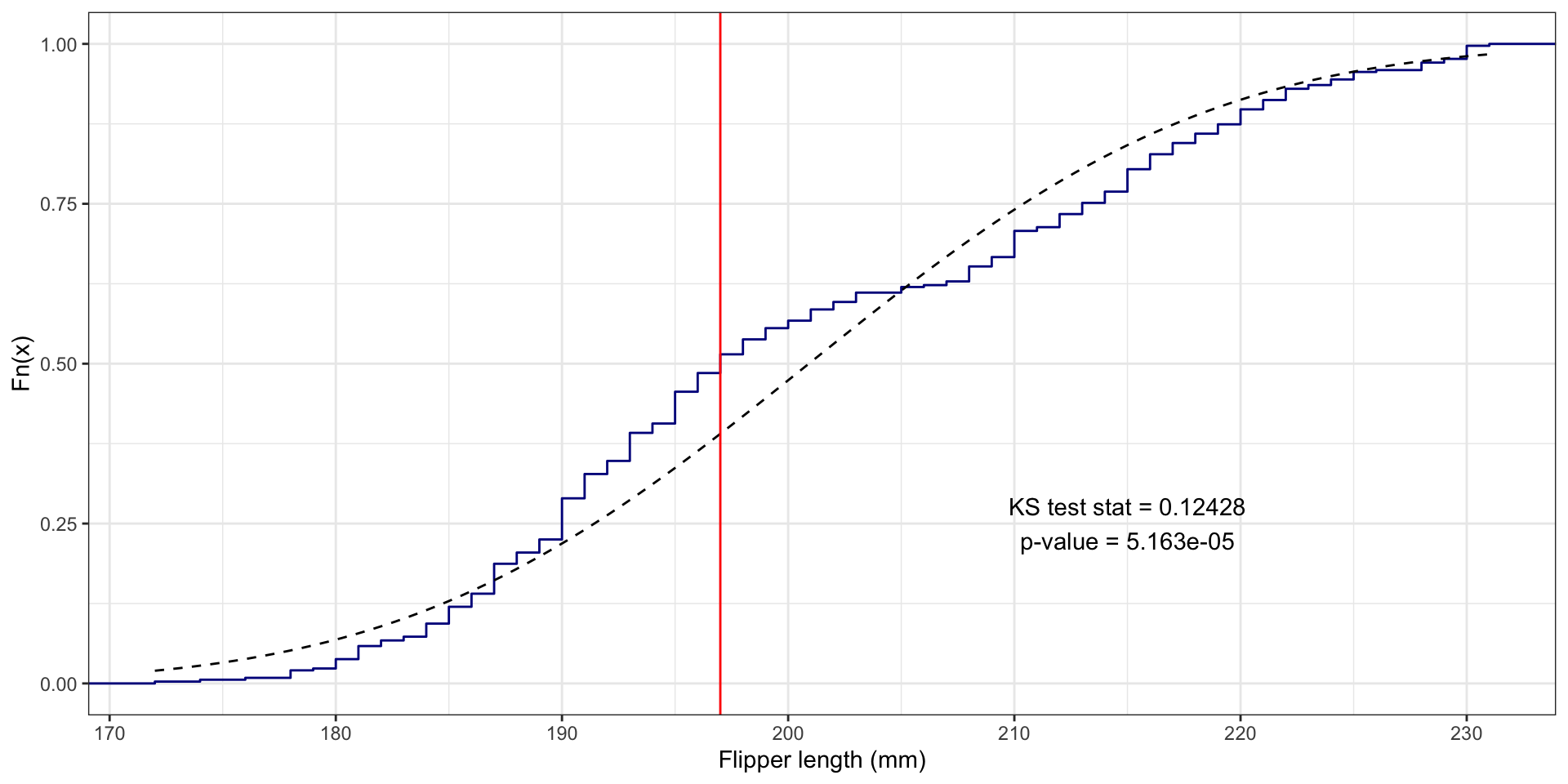
BONUS: Visualizing the KS test statistic
# First create the ECDF function for the variable:
fl_ecdf <- ecdf(penguins$flipper_length_mm)
# Compute the absolute value of the differences between the ECDF for the values
# and the theoretical values with assumed Normal distribution:
abs_ecdf_diffs <- abs(fl_ecdf(penguins$flipper_length_mm) - pnorm(penguins$flipper_length_mm,
mean = flipper_length_mean, sd = flipper_length_sd))
# Now find where the maximum difference is:
max_abs_ecdf_diff_i <- which.max(abs_ecdf_diffs)
# Get this flipper length value:
max_fl_diff_value <- penguins$flipper_length_mm[max_abs_ecdf_diff_i]
# Plot the ECDF with the theoretical Normal and KS test info:
penguins |>
ggplot(aes(x = flipper_length_mm)) +
stat_ecdf(color = "darkblue") +
# Use stat_function to draw the Normal ECDF
stat_function(fun = pnorm, args = list(mean = flipper_length_mean, sd = flipper_length_sd), color = "black", linetype = "dashed") +
# Draw KS test line:
geom_vline(xintercept = max_fl_diff_value, color = "red") +
# Add text with the test results (x and y are manually entered locations)
annotate(geom = "text", x = 215, y = .25, label = "KS test stat = 0.12428\np-value = 5.163e-05") +
labs(x = "Flipper length (mm)", y = "Fn(x)") + theme_bw()