# A tibble: 3 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
# ℹ 2 more variables: sex <fct>, year <int>Into High-Dimensional Data
2024-09-16
Reminders, previously, and today…
HW3 is due Wednesday!
HW4 is posted and due next Wednesday Sept 25th
Walked through visualiziations with scatterplots (always adjust the alpha!)
Displayed 2D joint distributions with contours, heatmaps, and hexagonal binning
Discussed approaches for visualizing conditional relationships
TODAY:
Into high-dimensional data
What type of structure do we want to capture?
Back to the penguins…
Pretend I give you this penguins dataset and I ask you to make a plot for every pairwise comparison…
We can create a pairs plot to see all pairwise relationships in one plot
Pairs plot can include the various kinds of pairwise plots we’ve seen:
Two quantitative variables: scatterplot
One categorical, one quantitative: side-by-side violins, stacked histograms, overlaid densities
Two categorical: stacked bars, side-by-side bars, mosaic plots
Create pairs plots with GGally
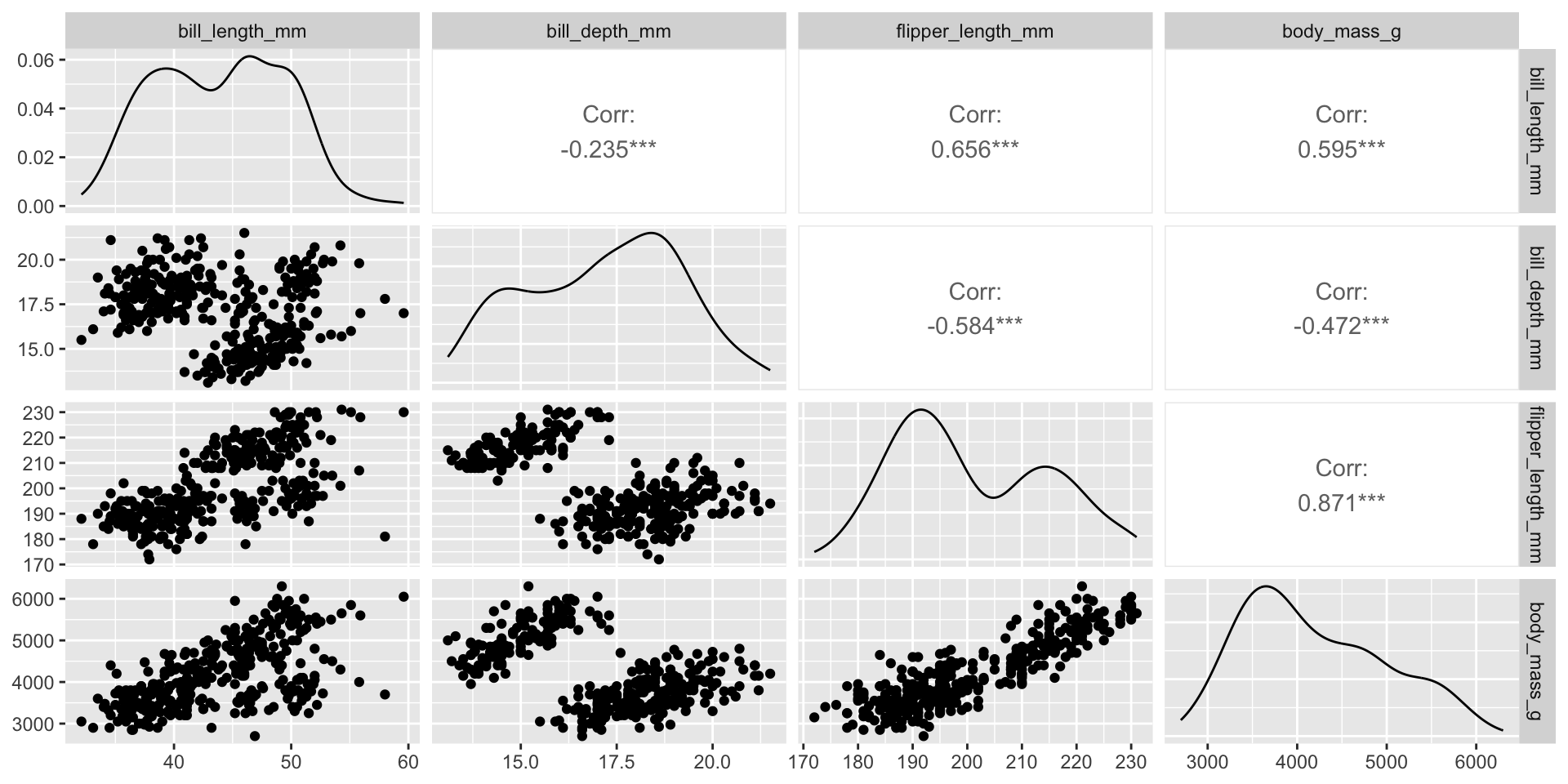
Create pairs plots with GGally
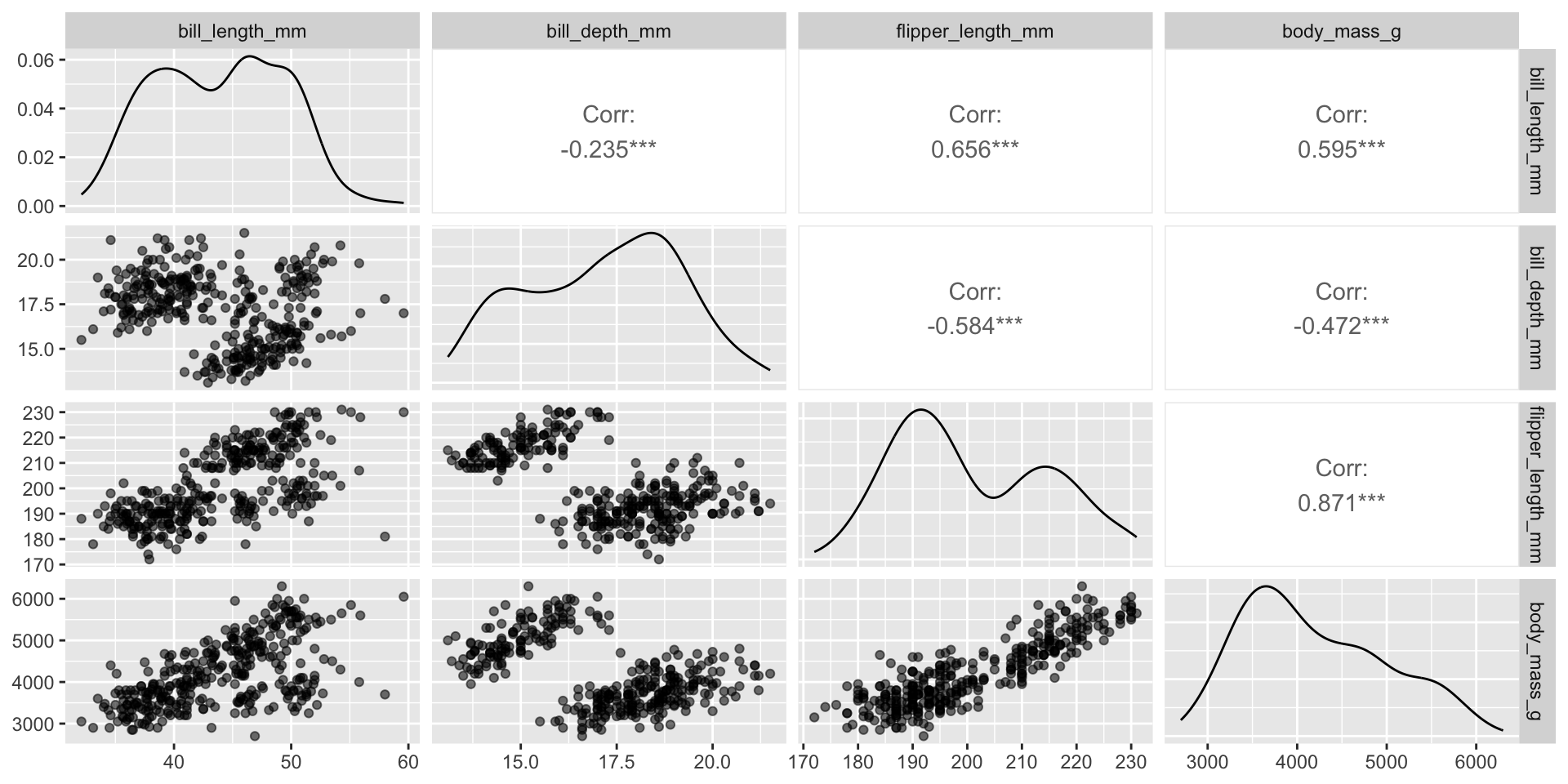
Flexibility in customization
Flexibility in customization
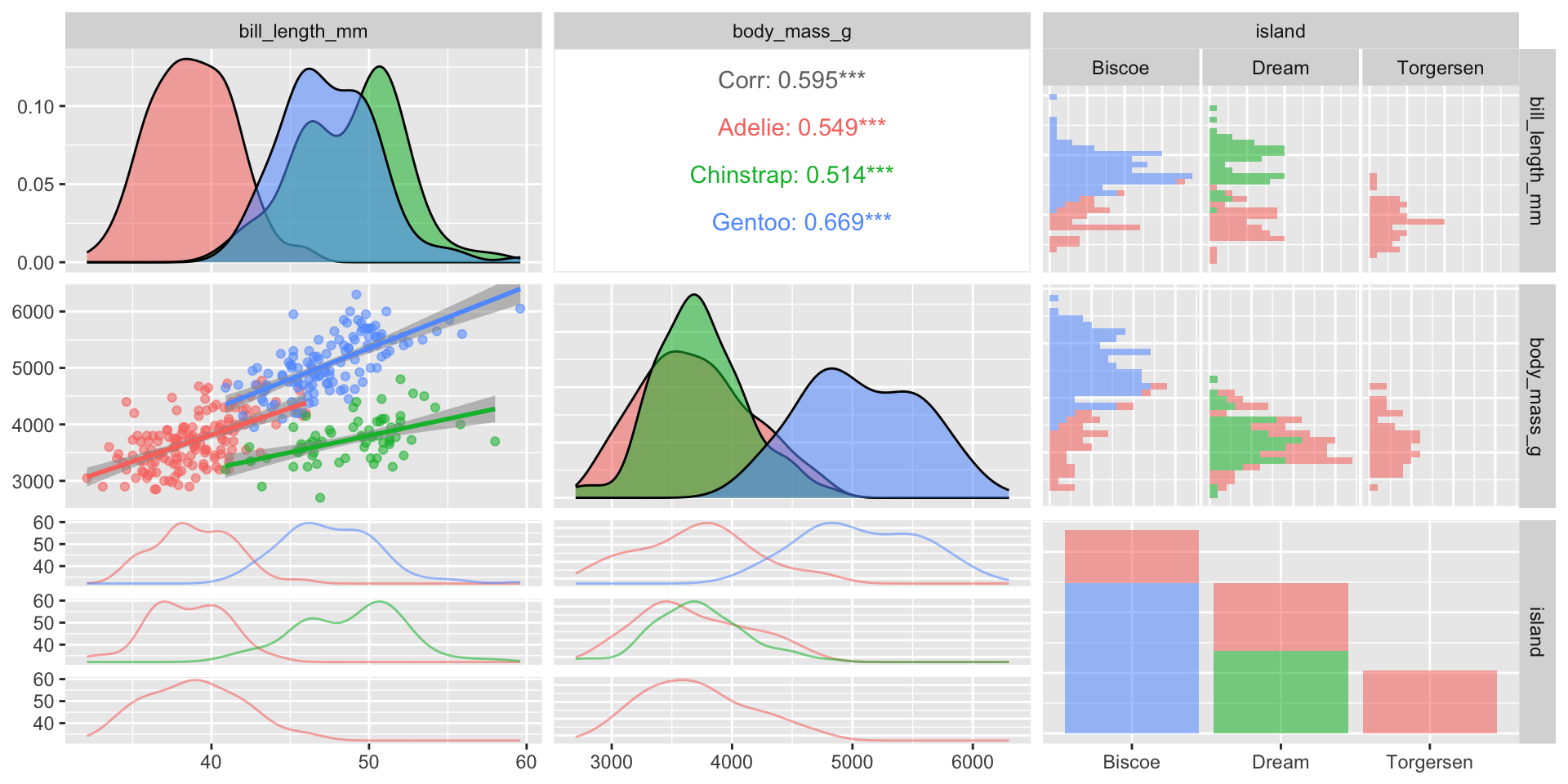
See Demo 03 for more!
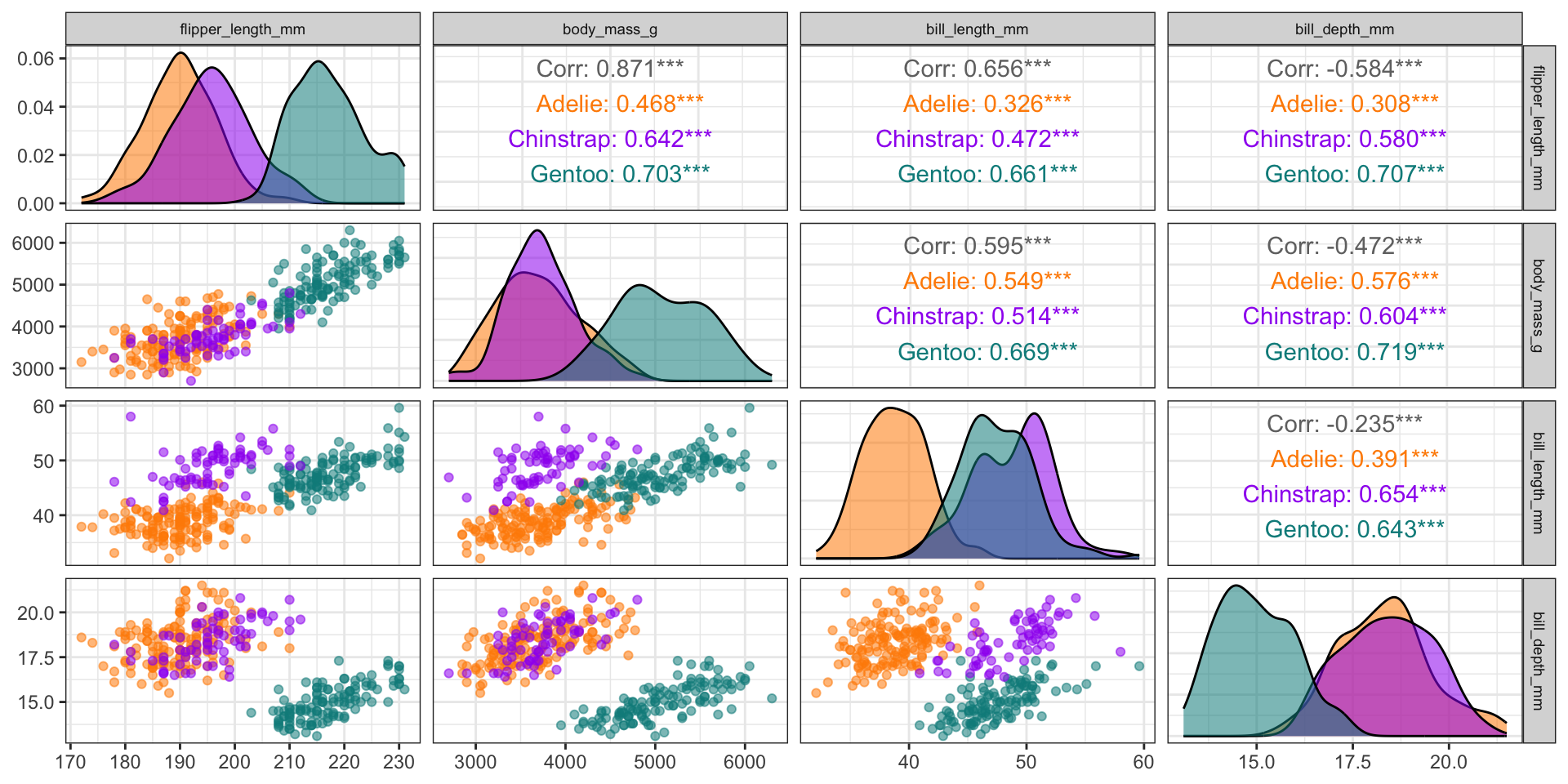
What about high-dimensional data?
Consider this dataset containing nutritional information about Starbucks drinks:
starbucks <-
read_csv("https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2021/2021-12-21/starbucks.csv") |>
# Convert columns to numeric that were saved as character
mutate(trans_fat_g = as.numeric(trans_fat_g), fiber_g = as.numeric(fiber_g))
starbucks |> slice(1)# A tibble: 1 × 15
product_name size milk whip serv_size_m_l calories total_fat_g
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 brewed coffee - dark roa… short 0 0 236 3 0.1
# ℹ 8 more variables: saturated_fat_g <dbl>, trans_fat_g <dbl>,
# cholesterol_mg <dbl>, sodium_mg <dbl>, total_carbs_g <dbl>, fiber_g <dbl>,
# sugar_g <dbl>, caffeine_mg <dbl>How do we visualize this dataset?
- Tedious task: make a series of pairs plots (one giant pairs plot would overwhelming)
What about high-dimensional data?
starbucks <-
read_csv("https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2021/2021-12-21/starbucks.csv") |>
# Convert columns to numeric that were saved as character
mutate(trans_fat_g = as.numeric(trans_fat_g), fiber_g = as.numeric(fiber_g))
starbucks |> slice(1)# A tibble: 1 × 15
product_name size milk whip serv_size_m_l calories total_fat_g
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 brewed coffee - dark roa… short 0 0 236 3 0.1
# ℹ 8 more variables: saturated_fat_g <dbl>, trans_fat_g <dbl>,
# cholesterol_mg <dbl>, sodium_mg <dbl>, total_carbs_g <dbl>, fiber_g <dbl>,
# sugar_g <dbl>, caffeine_mg <dbl>Goals to keep in mind with visualizing high-dimensional data:
Visualize structure among observations based on distances and projections (next lecture)
Visualize structure among variables using correlation as “distance”
Correlogram to visualize correlation matrix
Use the ggcorrplot package:
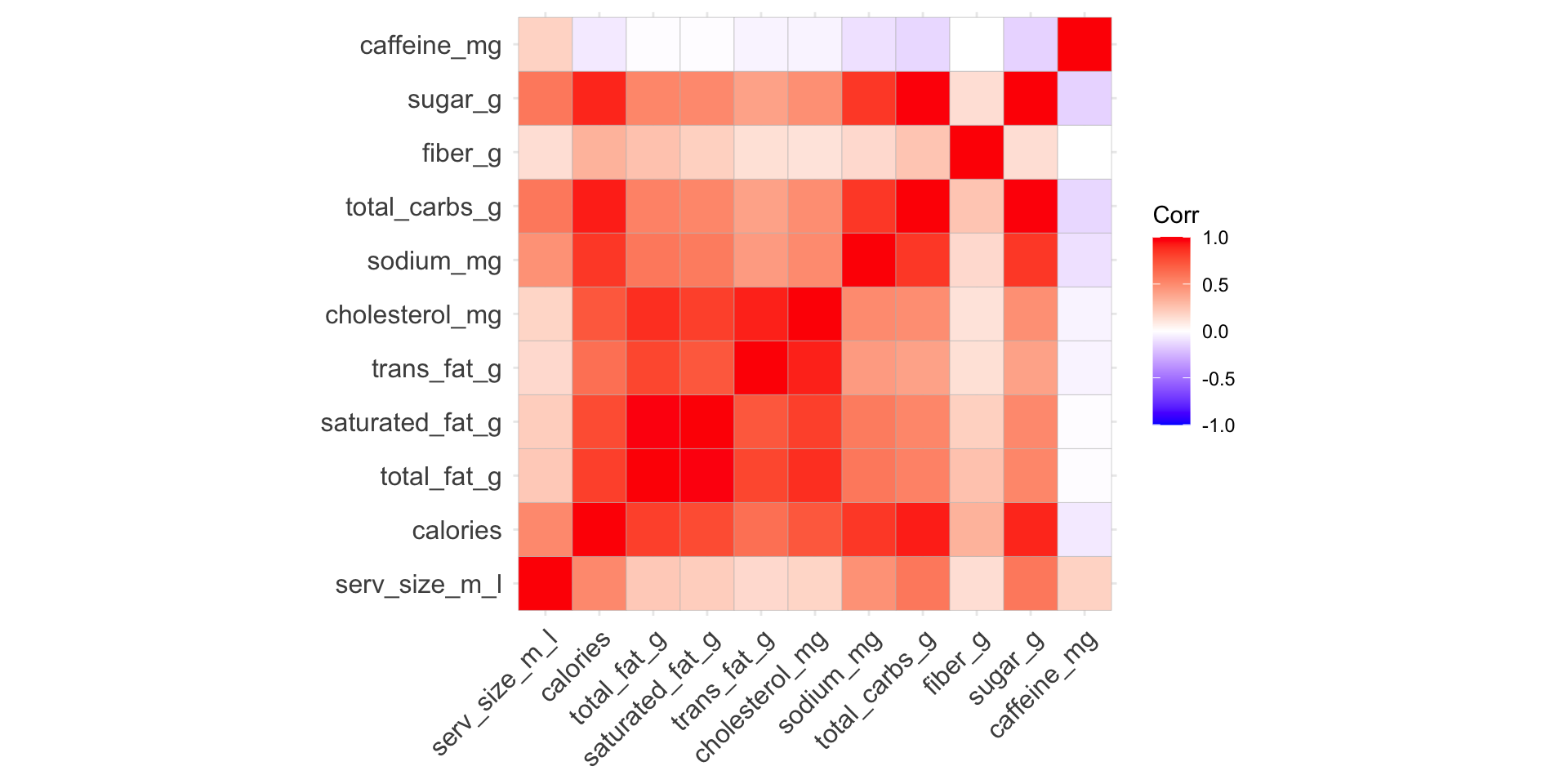
Options to customize correlogram
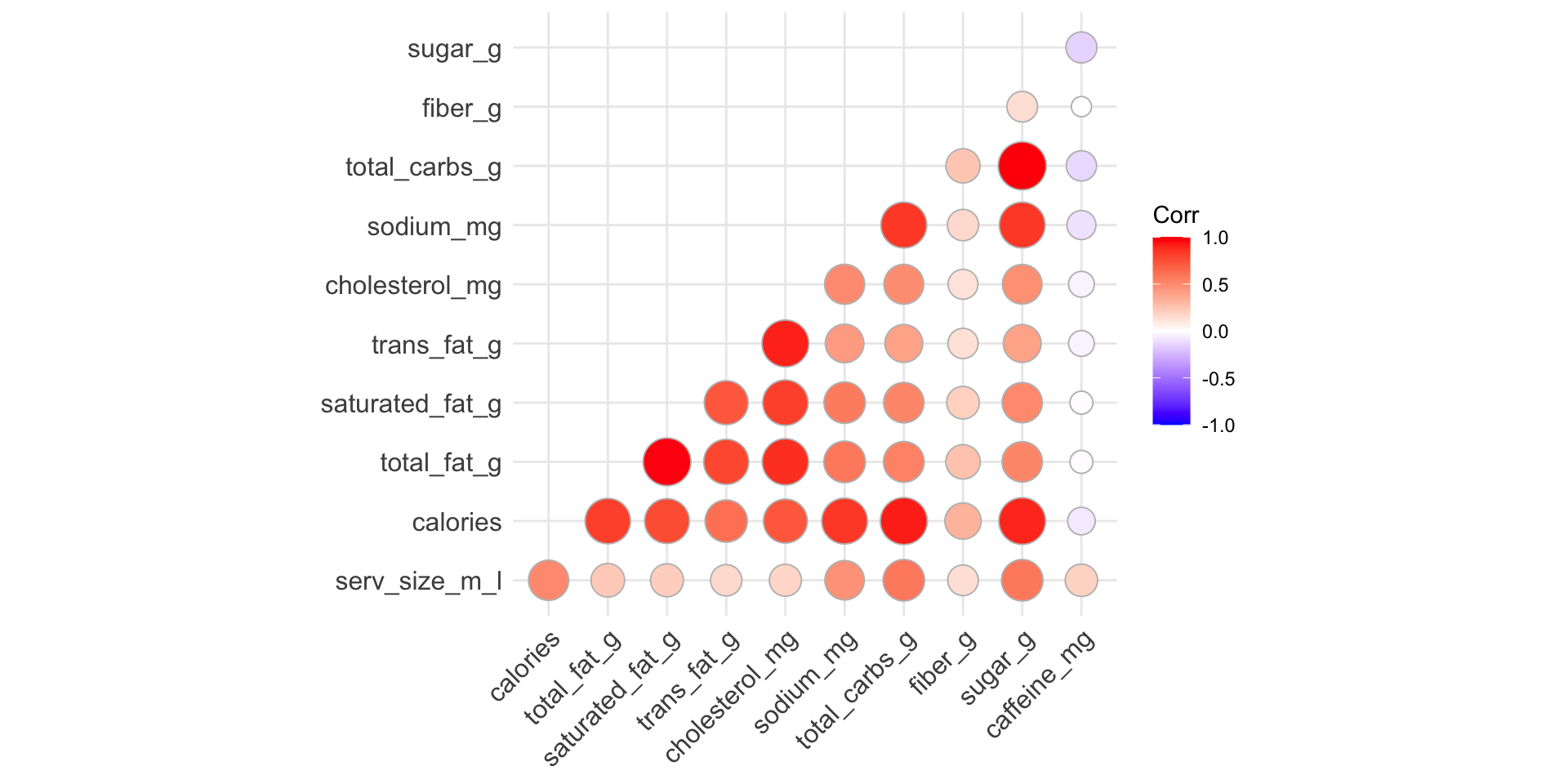
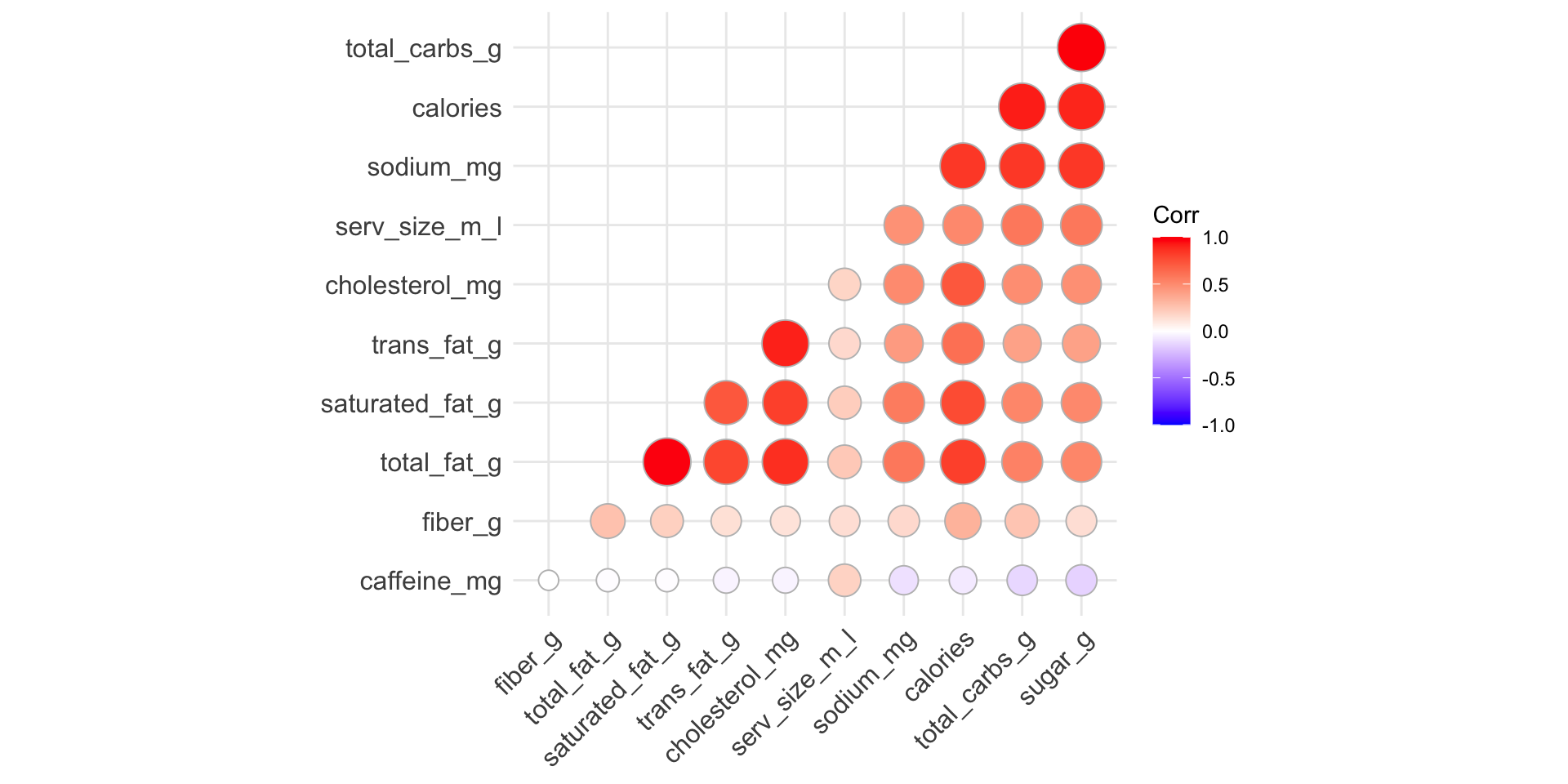
Reorder variables based on correlation
Heatmap displays of observations
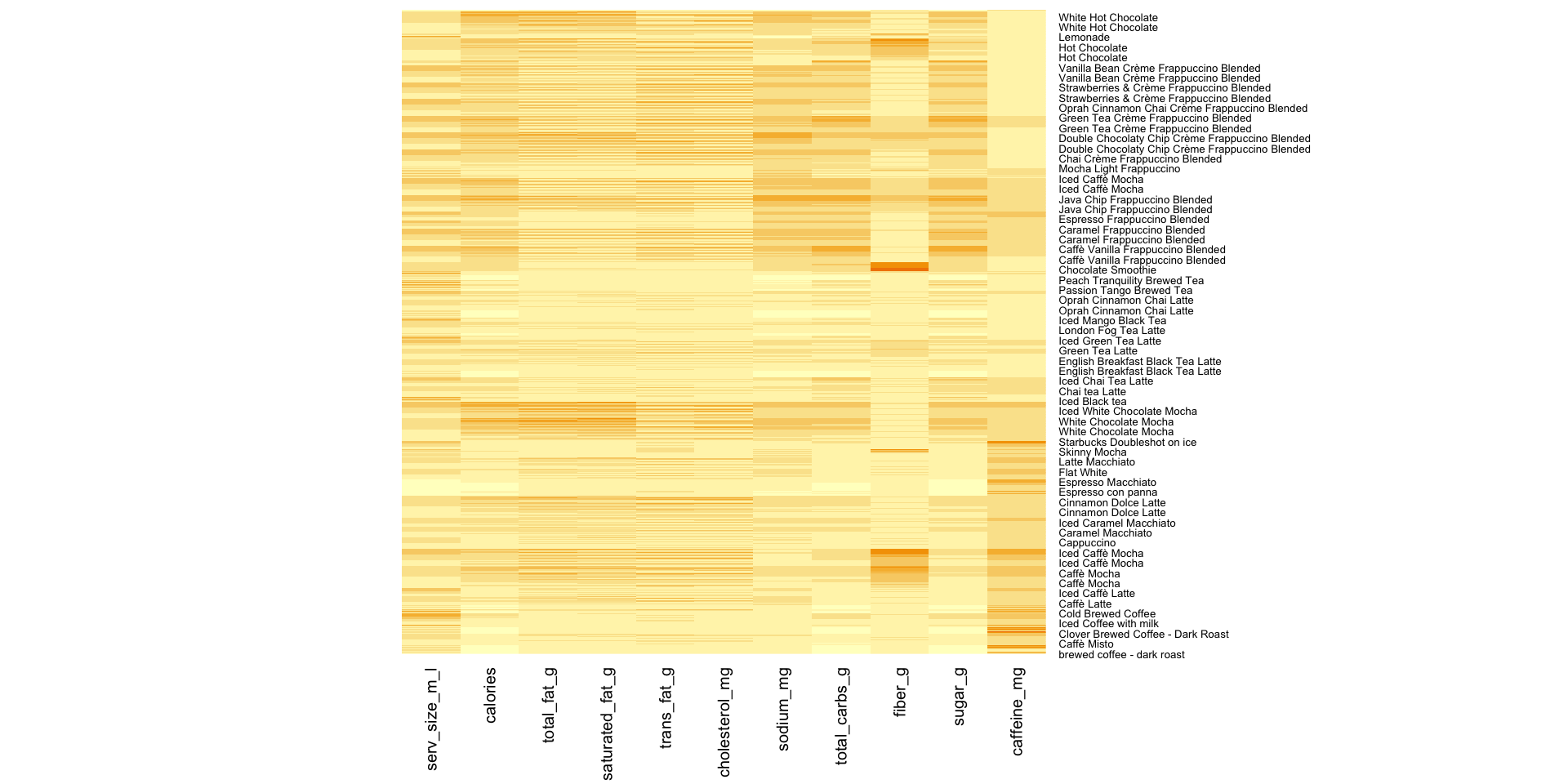
Manual version of heatmaps
starbucks |>
dplyr::select(product_name, serv_size_m_l:caffeine_mg) |>
pivot_longer(serv_size_m_l:caffeine_mg,
names_to = "variable",
values_to = "raw_value") |>
group_by(variable) |>
mutate(std_value = (raw_value - mean(raw_value)) / sd(raw_value)) |>
ungroup() |>
ggplot(aes(y = variable, x = product_name, fill = std_value)) +
geom_tile() +
theme_light() +
theme(axis.text.x = element_text(size = 1, angle = 45),
legend.position = "bottom") Manual version of heatmaps
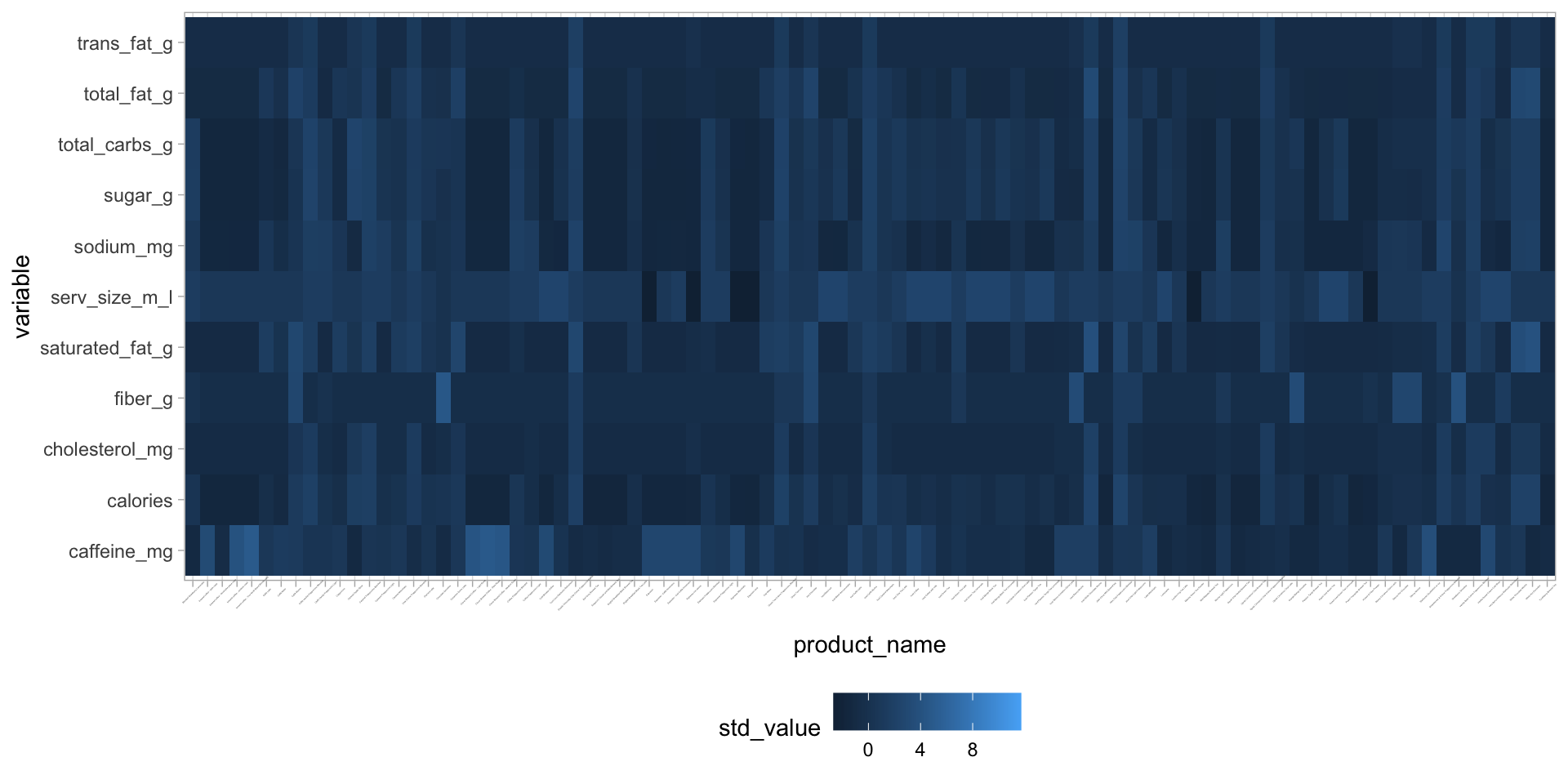
Manual version of heatmaps
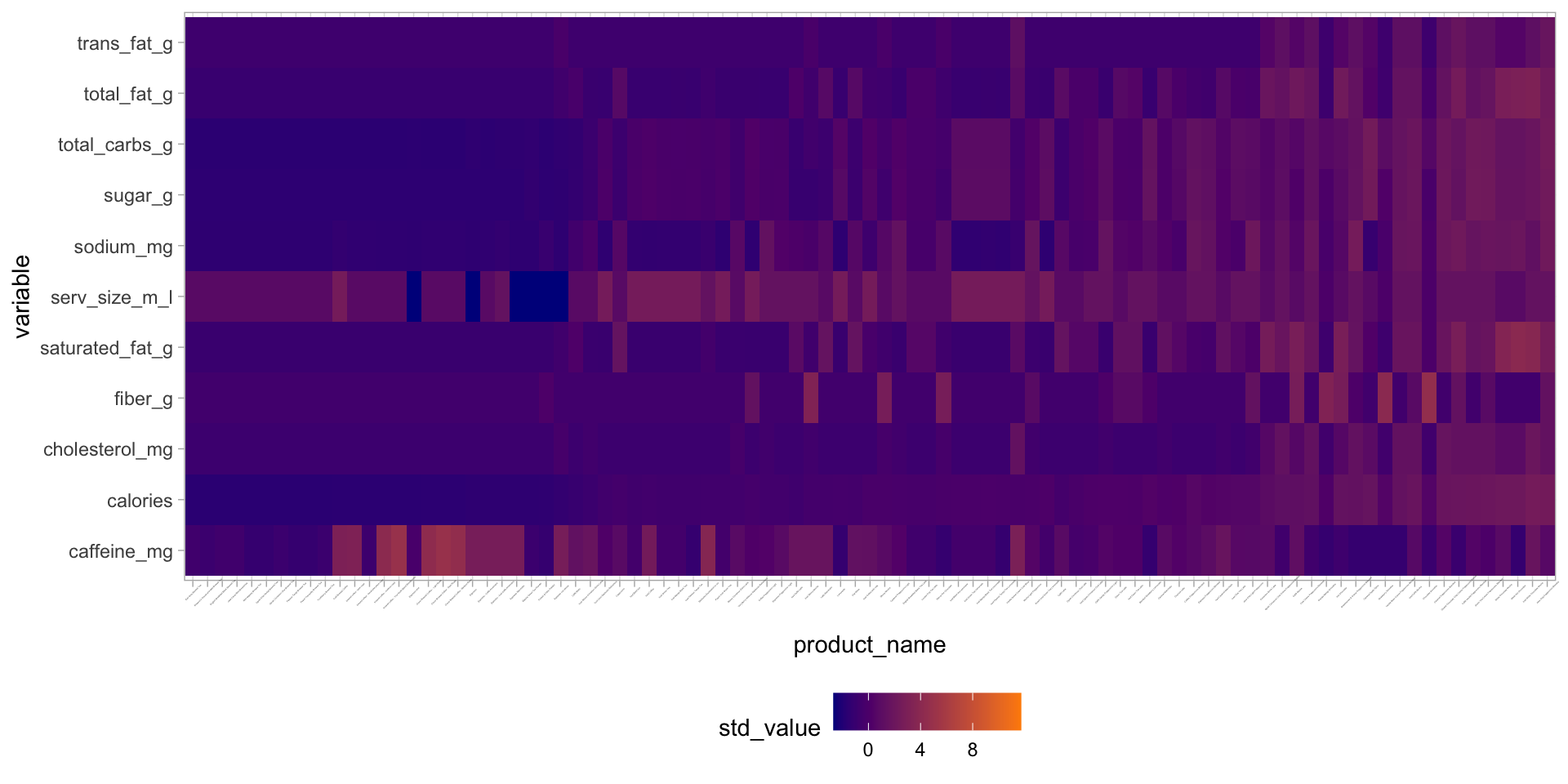
starbucks |>
dplyr::select(product_name, serv_size_m_l:caffeine_mg) |>
mutate(product_name = fct_reorder(product_name, calories)) |>
pivot_longer(serv_size_m_l:caffeine_mg,
names_to = "variable",
values_to = "raw_value") |>
group_by(variable) |>
mutate(std_value = (raw_value - mean(raw_value)) / sd(raw_value)) |>
ungroup() |>
ggplot(aes(y = variable, x = product_name, fill = std_value)) +
geom_tile() +
scale_fill_gradient(low = "darkblue", high = "darkorange") +
theme_light() +
theme(axis.text.x = element_text(size = 1, angle = 45),
legend.position = "bottom") Manual version of heatmaps
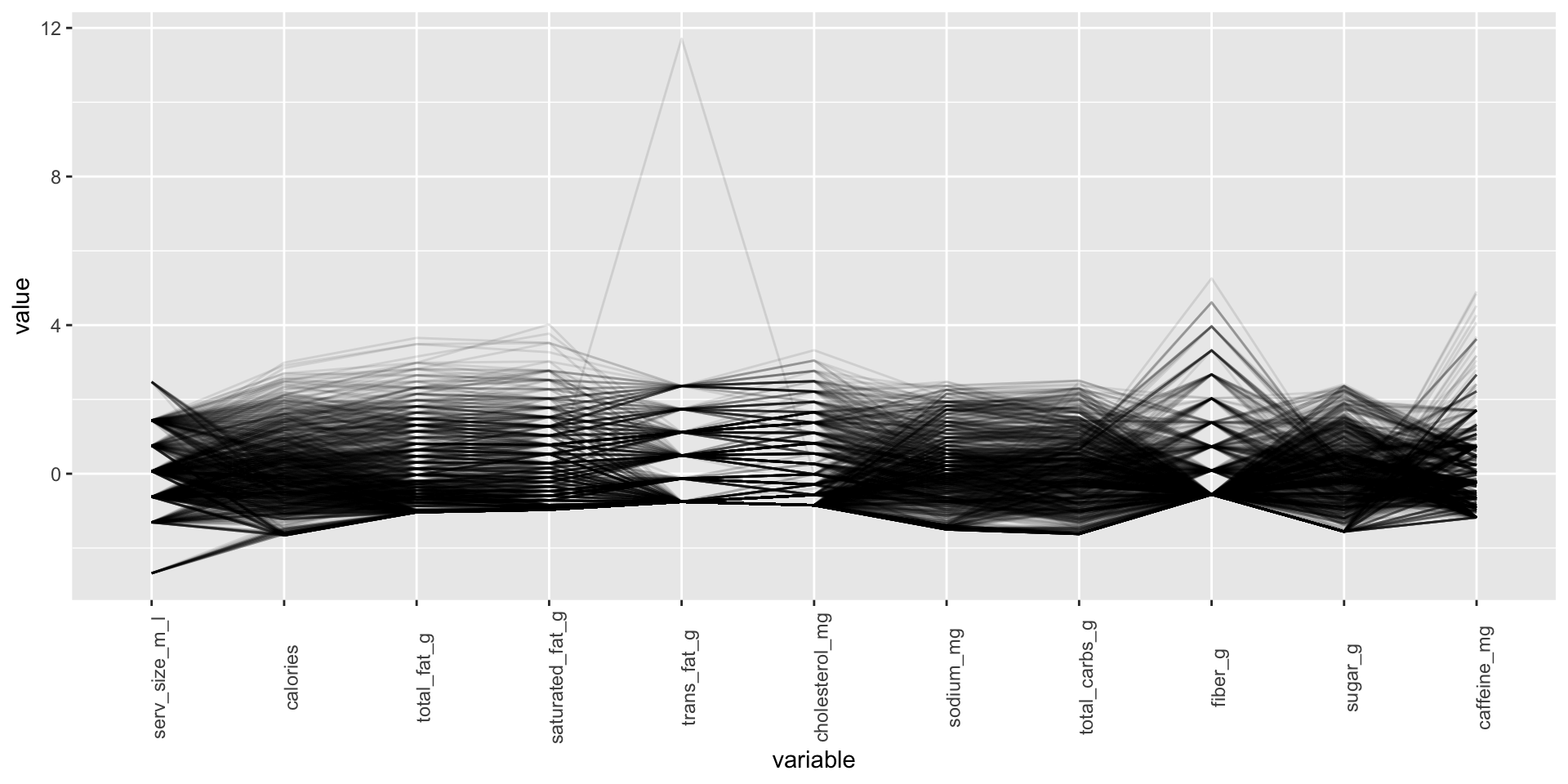
Parallel coordinates plot with ggparcoord
Recap and next steps
Discussed creating pairs plots for initial inspection of several variables
Began thinking about ways to displays dataset structure via correlations
Used heatmaps and parallel coordinates plot to capture observation and variable structure
HW3 is due Wednesday!
HW4 is posted due next Wednesday Sept 25th
Next time: More high-dimensional data
Recommended reading:
CW Chapter 12 Visualizing associations among two or more quantitative variables